Poster Session C
Imaging
Session: (1862–1894) Imaging of Rheumatic Diseases Poster II
1879: Artificial Intelligence for the Detection of Sacroiliitis on Magnetic Resonance Imaging in Patients with Axial Spondyloarthritis
Tuesday, November 14, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- SL
Seulkee Lee, MD, PhD (he/him/his)
Samsung Medical Center
Seoul, South KoreaDisclosure information not submitted.
Abstract Poster Presenter(s)
Seulkee Lee1, Uju Jeon2, Ji Hyun Lee3, Seonyoung Kang1, Hyungjin Kim1, Jaejoon Lee1, Myung Jin Chung2, Jinseok Kim4, Eun-Mi Koh5 and Hoon-Suk Cha1, 1Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea, 2Medical AI Research Center, Samsung Medical Center, Seoul, South Korea, 3Department of Radiology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, South Korea, 4Department of Rheumatology, Jeju National University Hospital, Jeju National University School of Medicine, Jeju-si, Jeju-do, South Korea, La Jolla, CA, South Korea, 5Health Insurance Review and Assessment Service, Seoul, South Korea
Background/Purpose: Magnetic resonance imaging (MRI) is important for the early detection of axial spondyloarthritis (axSpA). However, evaluation of sacroiliitis on MRI requires expertise because degenerative changes can mimic axSpA, and semiquantitative diagnosis remains subject to significant variation. Artificial intelligence (AI) has previously been applied to various MRI data including that of musculoskeletal system. Therefore, we developed an AI model for detecting in patients with axSpA, on MRI.
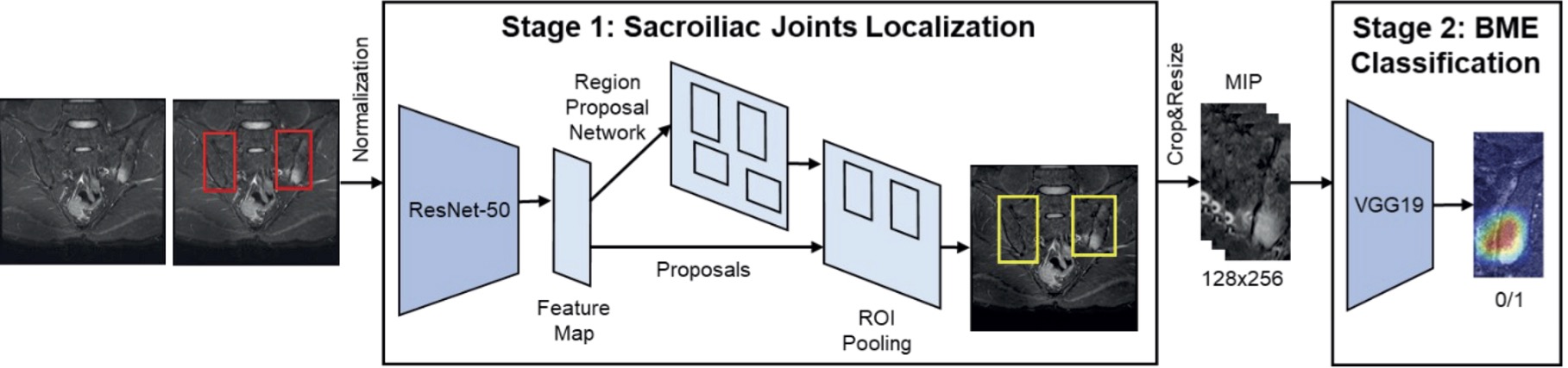
Methods: This study included MRI examinations of patients who underwent semi-coronal MRI scans of the sacroiliac joints owing to chronic back pain with short tau inversion recovery sequences between January 2010 and December 2021. Sacroiliitis was defined as a positive MRI finding according to the Assessment of Spondylo Arthritis international Society (ASAS) classification criteria for axSpA. The pipeline for the proposed automatic sacroiliitis classification method is shown in Figure 1. We developed a two-stage framework. First, the Faster R-CNN network extracted regions of interest (ROIs) to localize the sacroiliac joints. Maximum intensity projection (MIP) of three consecutive slices was used to mimic the reading of two adjacent slices. Second, the VGG-19 network determined the presence of sacroiliitis in localized ROIs. We augmented the positive dataset six-fold because of the smaller number of data points compared with the negative dataset. The sacroiliitis classification performance was measured using the accuracy and area under the receiver operating characteristic curve (AUROC). The prediction models were evaluated using three-round three-fold cross-validation.
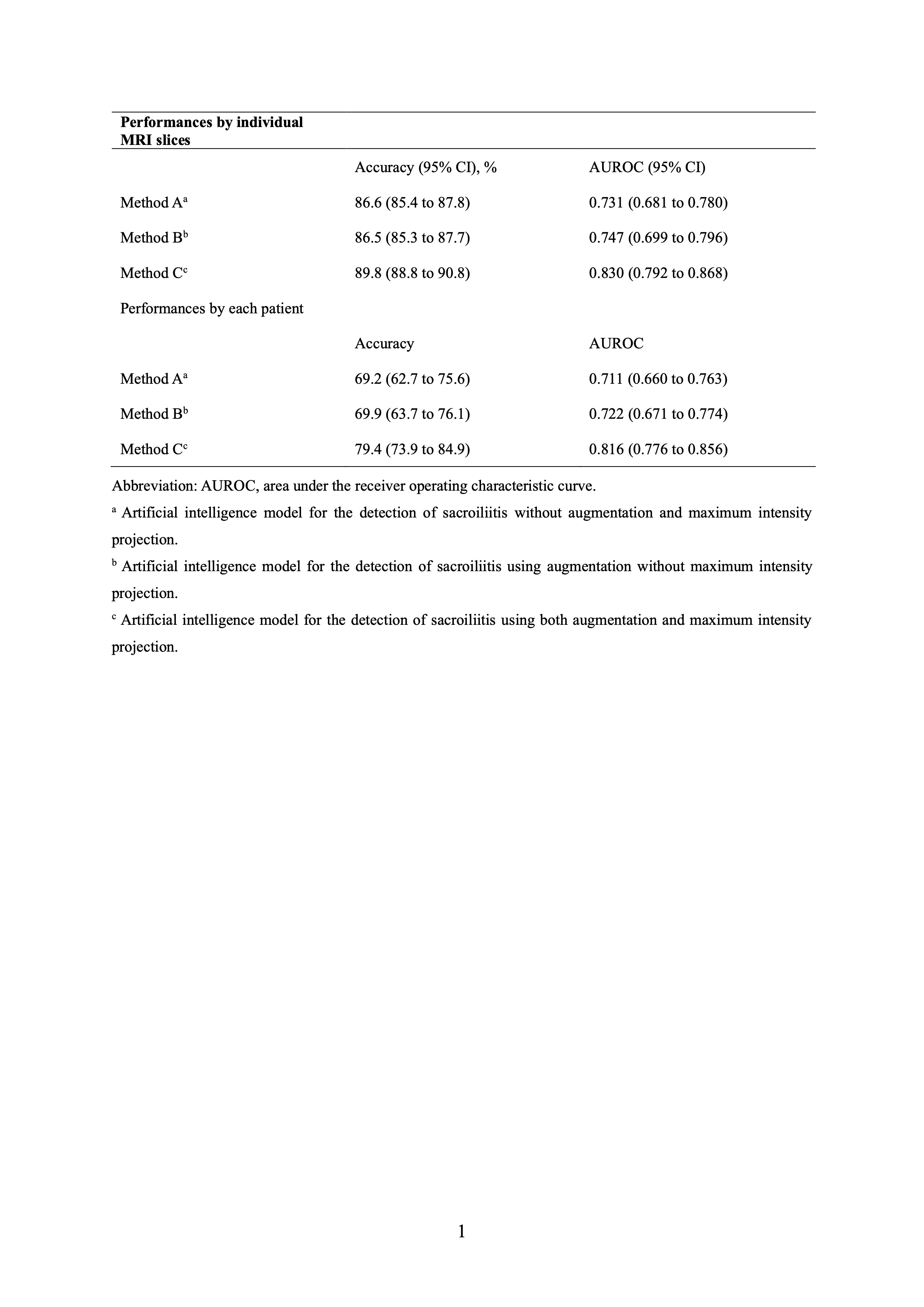
Results: A total of 296 participants with 4,746 MRI slices were included. Sacroiliitis was identified in 864 MRI slices from 119 participants. The mean accuracy and AUROC for the detection of sacroiliitis were 89.8% (95% CI, 88.8%–90.8%) and 0.830 (95% CI, 0.792–0.868), respectively, at the image level and 79.4% (95% CI, 73.9%–84.9%) and 0.816 (95% CI, 0.776–0.856), respectively, at the patient level. The confusion matrices for the detection of sacroiliitis per image and subject are shown in Figure 2. In the original model, without using MIP and dataset augmentation, the mean accuracy and AUROC were 86.6% (95% CI, 85.4%–87.8%) and 0.731 (95% CI, 0.681–0.780), respectively, at the image level and 69.2% (95% CI, 62.7%–75.6%) and 0.711 (95% CI, 0.660–0.763), respectively, at the individual level (Table 1). Improved performances were observed compared with the original model.
Conclusion: An AI model was developed for the detection of sacroiliitis using MRI, compatible with the ASAS criteria for axSpA, with the potential to aid MRI application in a wider clinical setting.
.jpg)
S. Lee: None; U. Jeon: None; J. Lee: None; S. Kang: None; H. Kim: None; J. Lee: None; M. Chung: None; J. Kim: None; E. Koh: None; H. Cha: None.
Background/Purpose: Magnetic resonance imaging (MRI) is important for the early detection of axial spondyloarthritis (axSpA). However, evaluation of sacroiliitis on MRI requires expertise because degenerative changes can mimic axSpA, and semiquantitative diagnosis remains subject to significant variation. Artificial intelligence (AI) has previously been applied to various MRI data including that of musculoskeletal system. Therefore, we developed an AI model for detecting in patients with axSpA, on MRI.
Methods: This study included MRI examinations of patients who underwent semi-coronal MRI scans of the sacroiliac joints owing to chronic back pain with short tau inversion recovery sequences between January 2010 and December 2021. Sacroiliitis was defined as a positive MRI finding according to the Assessment of Spondylo Arthritis international Society (ASAS) classification criteria for axSpA. The pipeline for the proposed automatic sacroiliitis classification method is shown in Figure 1. We developed a two-stage framework. First, the Faster R-CNN network extracted regions of interest (ROIs) to localize the sacroiliac joints. Maximum intensity projection (MIP) of three consecutive slices was used to mimic the reading of two adjacent slices. Second, the VGG-19 network determined the presence of sacroiliitis in localized ROIs. We augmented the positive dataset six-fold because of the smaller number of data points compared with the negative dataset. The sacroiliitis classification performance was measured using the accuracy and area under the receiver operating characteristic curve (AUROC). The prediction models were evaluated using three-round three-fold cross-validation.
Results: A total of 296 participants with 4,746 MRI slices were included. Sacroiliitis was identified in 864 MRI slices from 119 participants. The mean accuracy and AUROC for the detection of sacroiliitis were 89.8% (95% CI, 88.8%–90.8%) and 0.830 (95% CI, 0.792–0.868), respectively, at the image level and 79.4% (95% CI, 73.9%–84.9%) and 0.816 (95% CI, 0.776–0.856), respectively, at the patient level. The confusion matrices for the detection of sacroiliitis per image and subject are shown in Figure 2. In the original model, without using MIP and dataset augmentation, the mean accuracy and AUROC were 86.6% (95% CI, 85.4%–87.8%) and 0.731 (95% CI, 0.681–0.780), respectively, at the image level and 69.2% (95% CI, 62.7%–75.6%) and 0.711 (95% CI, 0.660–0.763), respectively, at the individual level (Table 1). Improved performances were observed compared with the original model.
Conclusion: An AI model was developed for the detection of sacroiliitis using MRI, compatible with the ASAS criteria for axSpA, with the potential to aid MRI application in a wider clinical setting.

Artificial intelligence framework to detect sacroiliitis in accordance with the Assessment of SpondyloArthritis International Society criteria for axial spondyloarthritis.
.jpg)
Confusion matrices of the first-round cross-validation using the proposed method (Method C) for detecting sacroiliitis (A) for individual MRI slices; (B) for each subject.

Performances of artificial intelligence models for the detection of sacroiliitis compatible with the Assessment of SpondyloArthritis international Society definition.
S. Lee: None; U. Jeon: None; J. Lee: None; S. Kang: None; H. Kim: None; J. Lee: None; M. Chung: None; J. Kim: None; E. Koh: None; H. Cha: None.



