Poster Session C
Spondyloarthritis (SpA) including psoriatic arthritis (PsA)
Session: (1776–1795) Spondyloarthritis Including Psoriatic Arthritis – Basic Science Poster
1791: Exploring the Mechanism of Anti-TNFα Therapy Non-response in Psoriatic Arthritis: The Role of TNF Receptor 2 Polymorphisms rs1061622
Tuesday, November 14, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall

James Sullivan, BA (he/him/his)
Cleveland Clinic Lerner College of Medicine
Cleveland Heights, OH, United StatesDisclosure information not submitted.
Abstract Poster Presenter(s)
James Sullivan1, Vandana Rai2, Jennifer Harvey2, Vincent Del Signore2, Unnikrishnan Chandrasekharan2 and M. Elaine Husni3, 1Cleveland Clinic Lerner College of Medicine, Cleveland, OH, 2Cleveland Clinic, Cleveland, OH, 3Cleveland Clinic / Department of Rheumatic and Immunologic Diseases, Cleveland, OH
Background/Purpose: Despite the widespread use of anti-TNFα therapy in psoriatic arthritis (PsA), a significant proportion of patients fail to achieve a complete treatment response. There are no predictive markers for response to TNFα blockade. Rs1061622 polymorphisms (T/T, T/G, or G/G genotypes) corresponds to a methionine with T allele or arginine with G allele at amino acid position 196 of TNFα receptor 2 (TNFR2). These polymorphisms in TNFR2 have been associated with treatment response in rheumatoid arthritis and PsA, with the G/G genotype being less likely to respond to anti-TNFα therapy compared to the T/T genotype. However, the underlying molecular mechanism explaining this association remains unknown. This study aimed to investigate the signaling differences between TNFR2 variants associated with rs1061622 polymorphisms carrying the T allele (TNFR2-196M) and G allele (TNFR2-196R), which could potentially elucidate the differential responsiveness to anti-TNFα therapy in PsA patients.
Methods: Gene expression studies were conducted in Jurkat T cells and primary human endothelial cells. Recombinant TNFR2-196M or TNFR2-196R were expressed in both cell types using a lentiviral approach. Cells were treated with TNFα alone (2 ng/mL) or TNFα in the presence of a TNFα neutralizing antibody. The expression of, ICAM-1, a TNFR2-dependent proinflammatory gene, was assessed using quantitative RT-PCR. These findings were further validated using human umbilical vein endothelial cells (HUVEC) with rs1061622 polymorphisms. The polymorphisms were determined by genotyping (T/T, T/G, and G/G) discarded umbilical cord tissues prior to isolating and culturing the endothelial cells. Signaling pathways through TNFR2 were assessed using RTqPCR and Western blot analysis.
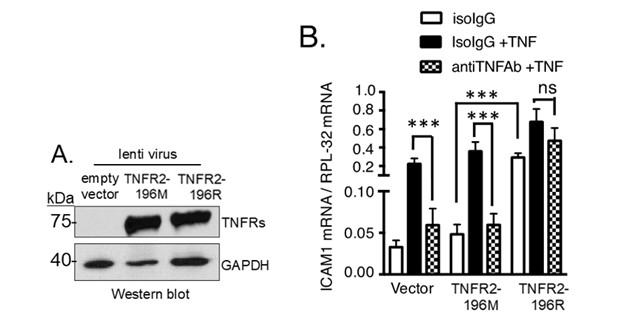
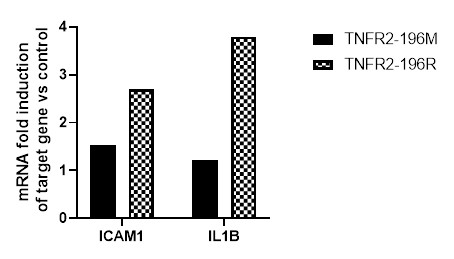
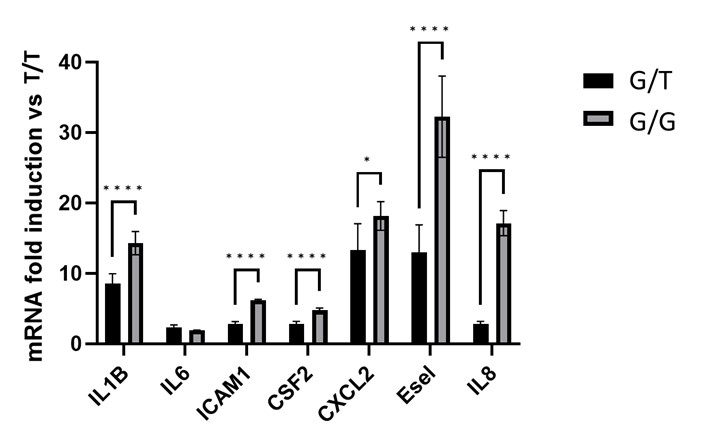
Results: In Jurkat T cells, successful expression of TNFR2-196M and TNFR2-196R was achieved (Figure 1A). Cells expressing TNFR2-196R exhibited increased basal expression of ICAM-1 compared to TNFR2-196M (Figure 1B). Importantly, treatment with a TNFα neutralizing antibody did not affect basal ICAM-1. Similarly, HUVEC cells over TNFR2-196R expressing (by lentiviral approach) showed increased ICAM-1 and IL-1β mRNA levels, while TNFR2-196M did not exhibit this effect (Figure 2). Furthermore, HUVEC isolated from subjects with G allele demonstrated higher basal expression of proinflammatory genes (IL-1β, IL-6, ICAM-1, GM-CSF2, CXCL2, E-selectin, Il-8) in the absence of TNFα (Figure 3). Notably, the TNFR2-independent gene P-selectin, did not show an increase in basal activity in cells expressing TNFR2-196R.
Conclusion: Our findings suggest that at least one G allele for the TNFR2 rs1061622 polymorphisms (TNFR-196R) confers a TNFα independent proinflammatory activity. These results may provide insights into a potential underlying mechanism for the association between rs1061622 polymorphisms and likelihood of response to anti-TNFα treatment in PsA. Further understanding of these signaling differences may contribute to the development of personalized treatment strategies for PsA patients based on their genetic profiles.
J. Sullivan: None; V. Rai: None; J. Harvey: None; V. Del Signore: None; U. Chandrasekharan: None; M. Husni: AbbVie, 1, 2, Amgen, 1, 2, Bristol-Myers Squibb, 1, 2, Eli Lilly, 1, 2, Janssen, 1, 2, Novartis, 1, 2, Pfizer, 1, 2, UCB, 1, 2.
Background/Purpose: Despite the widespread use of anti-TNFα therapy in psoriatic arthritis (PsA), a significant proportion of patients fail to achieve a complete treatment response. There are no predictive markers for response to TNFα blockade. Rs1061622 polymorphisms (T/T, T/G, or G/G genotypes) corresponds to a methionine with T allele or arginine with G allele at amino acid position 196 of TNFα receptor 2 (TNFR2). These polymorphisms in TNFR2 have been associated with treatment response in rheumatoid arthritis and PsA, with the G/G genotype being less likely to respond to anti-TNFα therapy compared to the T/T genotype. However, the underlying molecular mechanism explaining this association remains unknown. This study aimed to investigate the signaling differences between TNFR2 variants associated with rs1061622 polymorphisms carrying the T allele (TNFR2-196M) and G allele (TNFR2-196R), which could potentially elucidate the differential responsiveness to anti-TNFα therapy in PsA patients.
Methods: Gene expression studies were conducted in Jurkat T cells and primary human endothelial cells. Recombinant TNFR2-196M or TNFR2-196R were expressed in both cell types using a lentiviral approach. Cells were treated with TNFα alone (2 ng/mL) or TNFα in the presence of a TNFα neutralizing antibody. The expression of, ICAM-1, a TNFR2-dependent proinflammatory gene, was assessed using quantitative RT-PCR. These findings were further validated using human umbilical vein endothelial cells (HUVEC) with rs1061622 polymorphisms. The polymorphisms were determined by genotyping (T/T, T/G, and G/G) discarded umbilical cord tissues prior to isolating and culturing the endothelial cells. Signaling pathways through TNFR2 were assessed using RTqPCR and Western blot analysis.
Results: In Jurkat T cells, successful expression of TNFR2-196M and TNFR2-196R was achieved (Figure 1A). Cells expressing TNFR2-196R exhibited increased basal expression of ICAM-1 compared to TNFR2-196M (Figure 1B). Importantly, treatment with a TNFα neutralizing antibody did not affect basal ICAM-1. Similarly, HUVEC cells over TNFR2-196R expressing (by lentiviral approach) showed increased ICAM-1 and IL-1β mRNA levels, while TNFR2-196M did not exhibit this effect (Figure 2). Furthermore, HUVEC isolated from subjects with G allele demonstrated higher basal expression of proinflammatory genes (IL-1β, IL-6, ICAM-1, GM-CSF2, CXCL2, E-selectin, Il-8) in the absence of TNFα (Figure 3). Notably, the TNFR2-independent gene P-selectin, did not show an increase in basal activity in cells expressing TNFR2-196R.
Conclusion: Our findings suggest that at least one G allele for the TNFR2 rs1061622 polymorphisms (TNFR-196R) confers a TNFα independent proinflammatory activity. These results may provide insights into a potential underlying mechanism for the association between rs1061622 polymorphisms and likelihood of response to anti-TNFα treatment in PsA. Further understanding of these signaling differences may contribute to the development of personalized treatment strategies for PsA patients based on their genetic profiles.

Figure 1. TNFR2-196R expressing Jurkat T cells show increased expression of ICAM-1 in the absence of TNFα. A) Western blot showing TNFR2-196M or TNFR2-196R protein expression using lentiviral approach. B) Using RTqPCR basal ICAM-1 mRNA abundance is significantly increased in TNFR2-196R over-expressing cells using lentiviral approach compared to control (vector) or TNFR2-196M over-expressing cells via lentiviral approach (white bars). This constitutive TNFR2-196R activity is not inhibited after treatment with anti-TNFα monoclonal antibody.

Figure 2. TNFR2-196R expressing HUVEC show increased expression of ICAM-1 and IL-1β in the absence of TNFα. Using RTqPCR basal ICAM-1 and IL-1β mRNA abundance is significantly increased in TNFR2-196R over-expressing cells via lentiviral approach (checkered) compared to uninfected HUVEC (normalized) or TNFR2-196M over-expressing cells via lentiviral approach (grey bars). Results are the average of two independent experiments.

Figure 3. TNFα independent constitutive inflammatory activity in HUVEC expressing at least one G allele. Total RNA was extracted from HUVEC with T/T, T/G, or G/G genotypes. mRNA of indicated genes were quantified using RTqPCR after normalizing with mRNA levels of RPL-32, a housekeeping gene not induced by cytokines, including TNFα.
J. Sullivan: None; V. Rai: None; J. Harvey: None; V. Del Signore: None; U. Chandrasekharan: None; M. Husni: AbbVie, 1, 2, Amgen, 1, 2, Bristol-Myers Squibb, 1, 2, Eli Lilly, 1, 2, Janssen, 1, 2, Novartis, 1, 2, Pfizer, 1, 2, UCB, 1, 2.



