Poster Session B
Epidemiology, health policy and outcomes
Session: (0965–0992) Epidemiology & Public Health Poster II
0985: Assessing the Value of Comorbidity Clusters in Predicting Clinical Outcomes in Rheumatoid Arthritis: A Machine Learning Approach Using a Very Large US Registry
Monday, November 13, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
.png)
Daniel H. Solomon, MD, MPH
Brigham and Women's Hospital
Newton, MA, United StatesDisclosure(s): Janssen: Grant/Research Support (Ongoing)
Abstract Poster Presenter(s)
Daniel Solomon1, Fredrik Johansson2, Hongshu Guan3, Leah Santacroce4, Lin Guo5, Wendi Malley5 and Heather Litman5, 1Brigham and Women's Hospital, Newton, MA, 2Chalmers University of Technology, Goteborg, Sweden, 3Brigham and Women's Hospital, Boston, MA, 4Brigham and Women's Hospital, Boston, MA, 5CorEvitas, LLC, Waltham, MA
Background/Purpose: Comorbid conditions are very common in rheumatoid arthritis (RA) and several prior studies have derived comorbidity clusters using machine learning (ML). Clustering using ML is straightforward, but clusters only have value if they better explain clinical outcomes. We applied various ML algorithms to compare the clusters of comorbidities derived and to assess the value of the clusters for predicting clinical disease activity (CDAI) and function.
Methods: A large US-based RA registry, CorEvitas, was used to identify patients for the analysis. We assessed the presence of 24 comorbidities, and ML was used to derive comorbidity clusters. K-mode, K-mean, regression-based, and hierarchical clustering was used. To assess the value of the clusters, we compared them in clinical outcome models predicting clinical disease activity index (CDAI) and health assessment questionnaire (HAQ). We used data from the first three years of the six-year study period to derive clusters and assess time-averaged values for CDAI and HAQ during the latter three years. Model fit was assessed via adjusted R2 and Root Mean Square Error for a series of models that included clusters from K-mode and each of the 24 comorbidities separately. K-mode was selected as it was representative of the ML-based clustering algorithms.
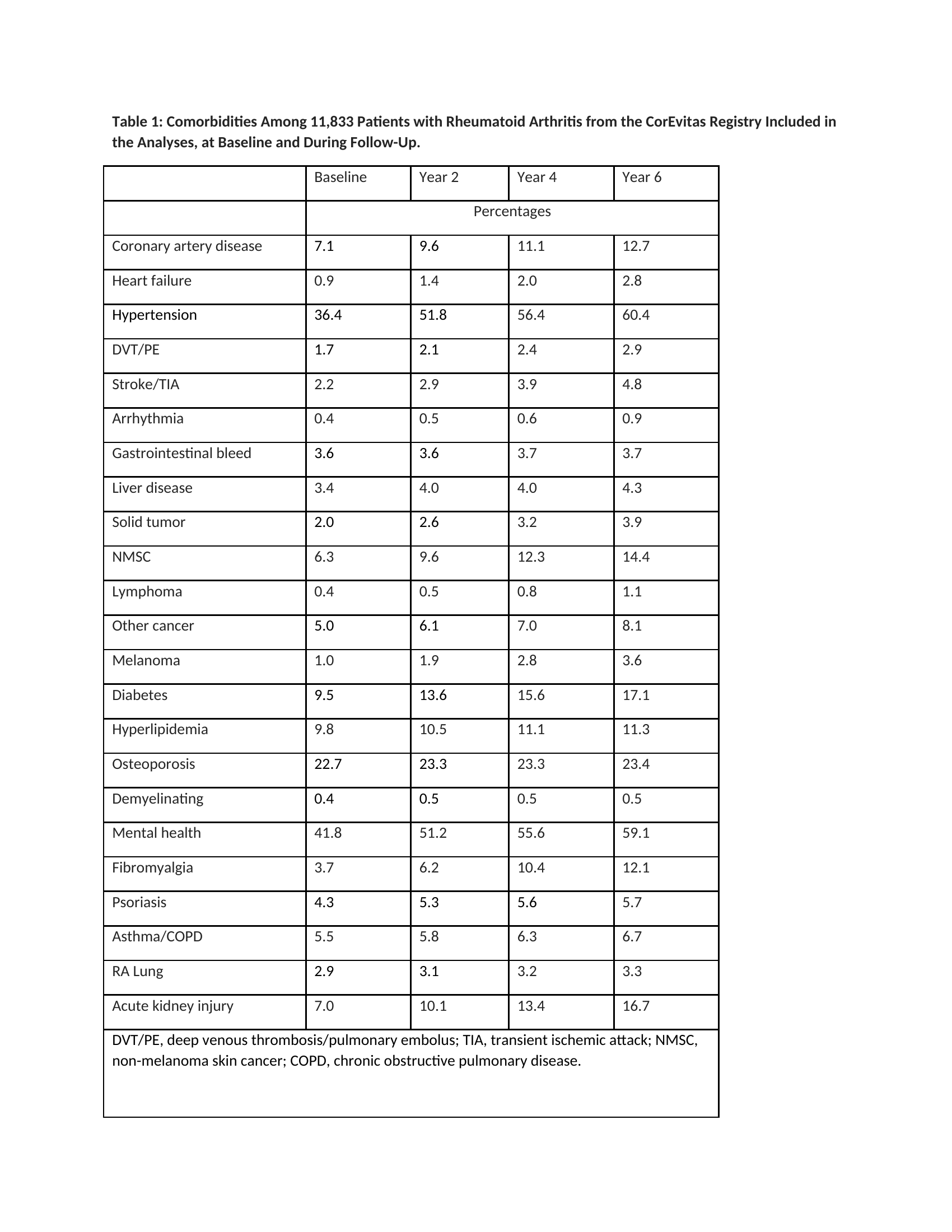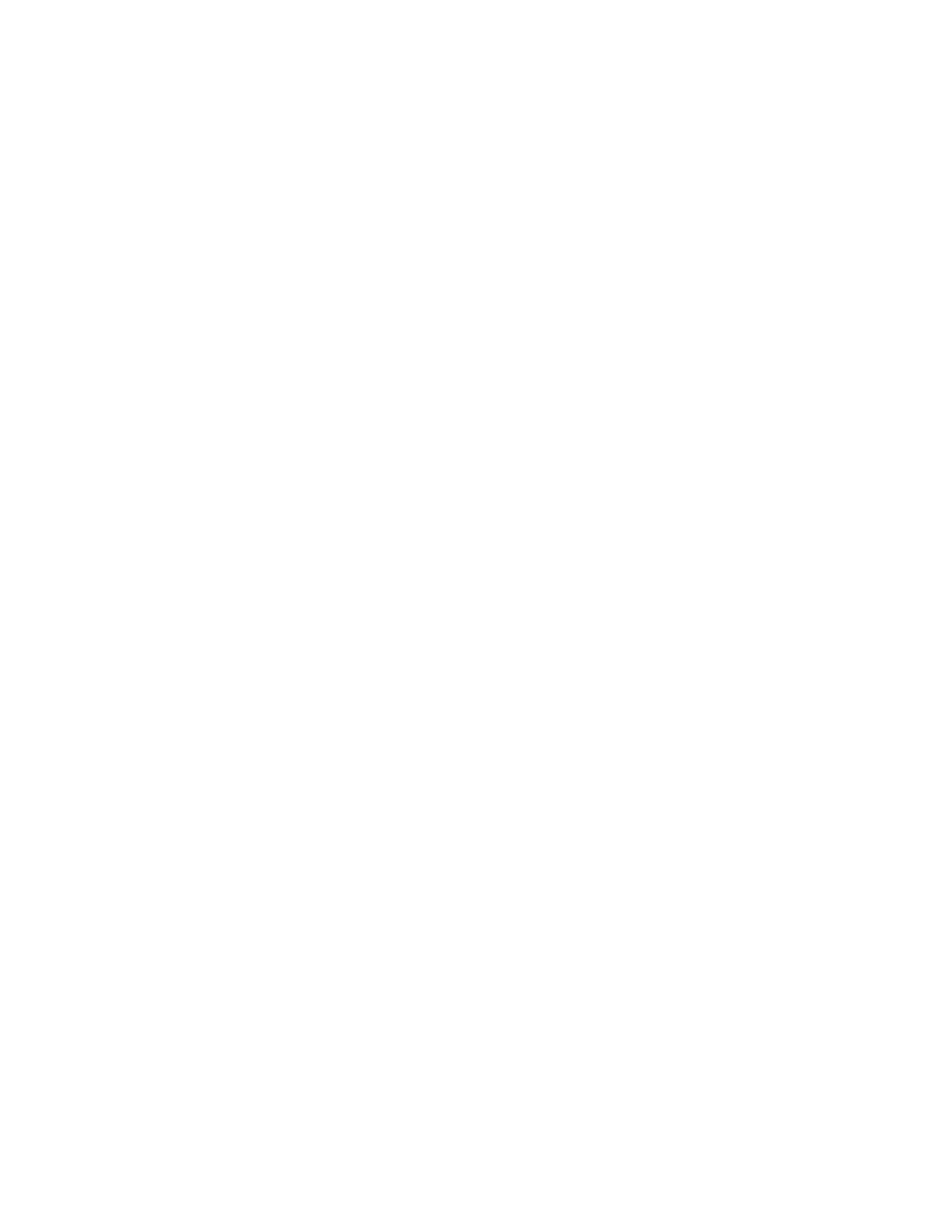
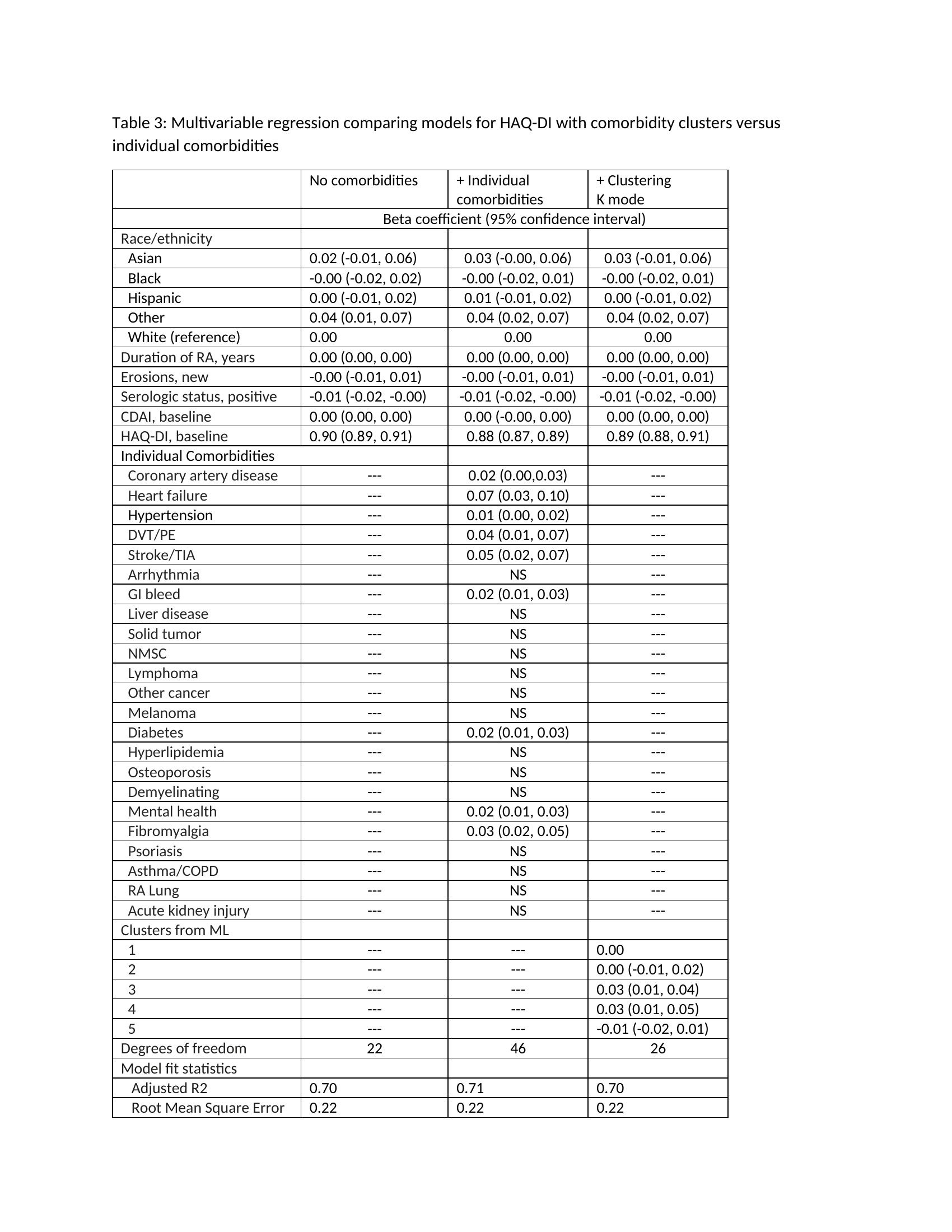
Results: 11,883 patients with RA were included who had longitudinal data over 6 years. At baseline, patients were on average 59 (SD 8) years of age, 77% were women, CDAI was 11.1 (SD 3.4, moderate disease activity), HAQ was 0.32 (SD 0.11), and disease duration was 10.9 (SD 4.3) years. During the six years of follow-up, the percentage of patients with various comorbidities increased (Table 1). Using five clusters produced by the K-mode ML algorithm, multivariable regression models with time-averaged CDAI as an outcome found that entering K-mode comorbidity clusters produced similarly strong models as models with each of the 24 separate comorbidities entered individually (Table 2). The same patterns were observed for HAQ (Table3). The other ML-based clustering algorithms produced very similar model results.
Conclusion: Clustering comorbidities using ML algorithms is not computationally complex but often results in clusters that are difficult to interpret from a clinical standpoint. While ML clustering is very useful for biologic modeling, using clusters to predict outcomes produces models with similar fit as those with individual comorbidities. Other use cases for comorbidity clusters might help demonstrate underlying biology.
.gif)
D. Solomon: CorEvitas, 5, Janssen, 5, Moderna, 5, Novartis, 5; F. Johansson: None; H. Guan: None; L. Santacroce: None; L. Guo: CorEvitas, LLC, 3; W. Malley: None; H. Litman: CorEvitas, 3, 12, Shareholder.
Background/Purpose: Comorbid conditions are very common in rheumatoid arthritis (RA) and several prior studies have derived comorbidity clusters using machine learning (ML). Clustering using ML is straightforward, but clusters only have value if they better explain clinical outcomes. We applied various ML algorithms to compare the clusters of comorbidities derived and to assess the value of the clusters for predicting clinical disease activity (CDAI) and function.
Methods: A large US-based RA registry, CorEvitas, was used to identify patients for the analysis. We assessed the presence of 24 comorbidities, and ML was used to derive comorbidity clusters. K-mode, K-mean, regression-based, and hierarchical clustering was used. To assess the value of the clusters, we compared them in clinical outcome models predicting clinical disease activity index (CDAI) and health assessment questionnaire (HAQ). We used data from the first three years of the six-year study period to derive clusters and assess time-averaged values for CDAI and HAQ during the latter three years. Model fit was assessed via adjusted R2 and Root Mean Square Error for a series of models that included clusters from K-mode and each of the 24 comorbidities separately. K-mode was selected as it was representative of the ML-based clustering algorithms.
Results: 11,883 patients with RA were included who had longitudinal data over 6 years. At baseline, patients were on average 59 (SD 8) years of age, 77% were women, CDAI was 11.1 (SD 3.4, moderate disease activity), HAQ was 0.32 (SD 0.11), and disease duration was 10.9 (SD 4.3) years. During the six years of follow-up, the percentage of patients with various comorbidities increased (Table 1). Using five clusters produced by the K-mode ML algorithm, multivariable regression models with time-averaged CDAI as an outcome found that entering K-mode comorbidity clusters produced similarly strong models as models with each of the 24 separate comorbidities entered individually (Table 2). The same patterns were observed for HAQ (Table3). The other ML-based clustering algorithms produced very similar model results.
Conclusion: Clustering comorbidities using ML algorithms is not computationally complex but often results in clusters that are difficult to interpret from a clinical standpoint. While ML clustering is very useful for biologic modeling, using clusters to predict outcomes produces models with similar fit as those with individual comorbidities. Other use cases for comorbidity clusters might help demonstrate underlying biology.

.gif)

D. Solomon: CorEvitas, 5, Janssen, 5, Moderna, 5, Novartis, 5; F. Johansson: None; H. Guan: None; L. Santacroce: None; L. Guo: CorEvitas, LLC, 3; W. Malley: None; H. Litman: CorEvitas, 3, 12, Shareholder.



