Poster Session B
Rheumatoid arthritis (RA)
Session: (1264–1307) RA – Diagnosis, Manifestations, and Outcomes Poster II
1290: Development of a Genetic Risk Score for Pain in Rheumatoid Arthritis
Monday, November 13, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- KM
Katie McMenamin, MD, BS (she/her/hers)
Boston University
Boston, MA, United StatesDisclosure information not submitted.
Abstract Poster Presenter(s)
Katie McMenamin1, Kristin Wipfler2, Austin Wheeler3, Grant Cannon4, K Wysham5, Brian Sauer6, Bryant England3, Kaleb Michaud3, Ted R Mikuls7 and Joshua Baker8, 1Boston University, Boston, MA, 2FORWARD, The National Databank for Rheumatic Diseases, Omaha, NE, 3University of Nebraska Medical Center, Omaha, NE, 4University of Utah and Salt Lake City VA, Salt Lake City, UT, 5VA Puget Sound/University of Washington, Seattle, WA, 6Salt Lake City VA/University of Utah, Salt Lake City, UT, 7Division of Rheumatology and Immunology, University of Nebraska Medical Center, Omaha, NE, 8University of Pennsylvania, Philadelphia, PA
Background/Purpose: Pain in rheumatoid arthritis (RA) is multifactorial and involves processes beyond inflammation such as peripheral and central pain processing. Several genes have been associated with pain processing through studies of fibromyalgia (FM), but it is largely unknown how genetics may affect pain in other rheumatic diseases. The aim of this study was to determine if single nucleotide polymorphisms (SNPs) associated with FM also have an impact on pain and disease activity in patients with RA.
Methods: Participants with RA with whole genome SNP data were included from two independent cohorts: 1) FORWARD (National Data Bank for Rheumatic Diseases) and 2) Veterans Affairs RA registry (VARA). Linear regression was used to determine the relationship between 30 individual SNPs previously associated with FM (Jannsen et al. An Acad Bras Ciênc. 2021) and baseline pain scores in FORWARD adjusting for age, sex and race. A genetic risk score (GRS) was generated using all 30 SNPs, weighted by the coefficients for each SNP from this regression. Linear regression was then used to assess the relationship between the GRS and cross-sectional (baseline) pain (visual analogue scale, range 0-10) in FORWARD (training dataset) and VARA (validation dataset). Similarly, the association of GRS with pain during longitudinal follow-up was determined using regression with generalized estimating equations adjusting for age, sex and race. In addition, the associations between GRS and disease activity, at enrollment and during longitudinal follow-up in each study were determined.
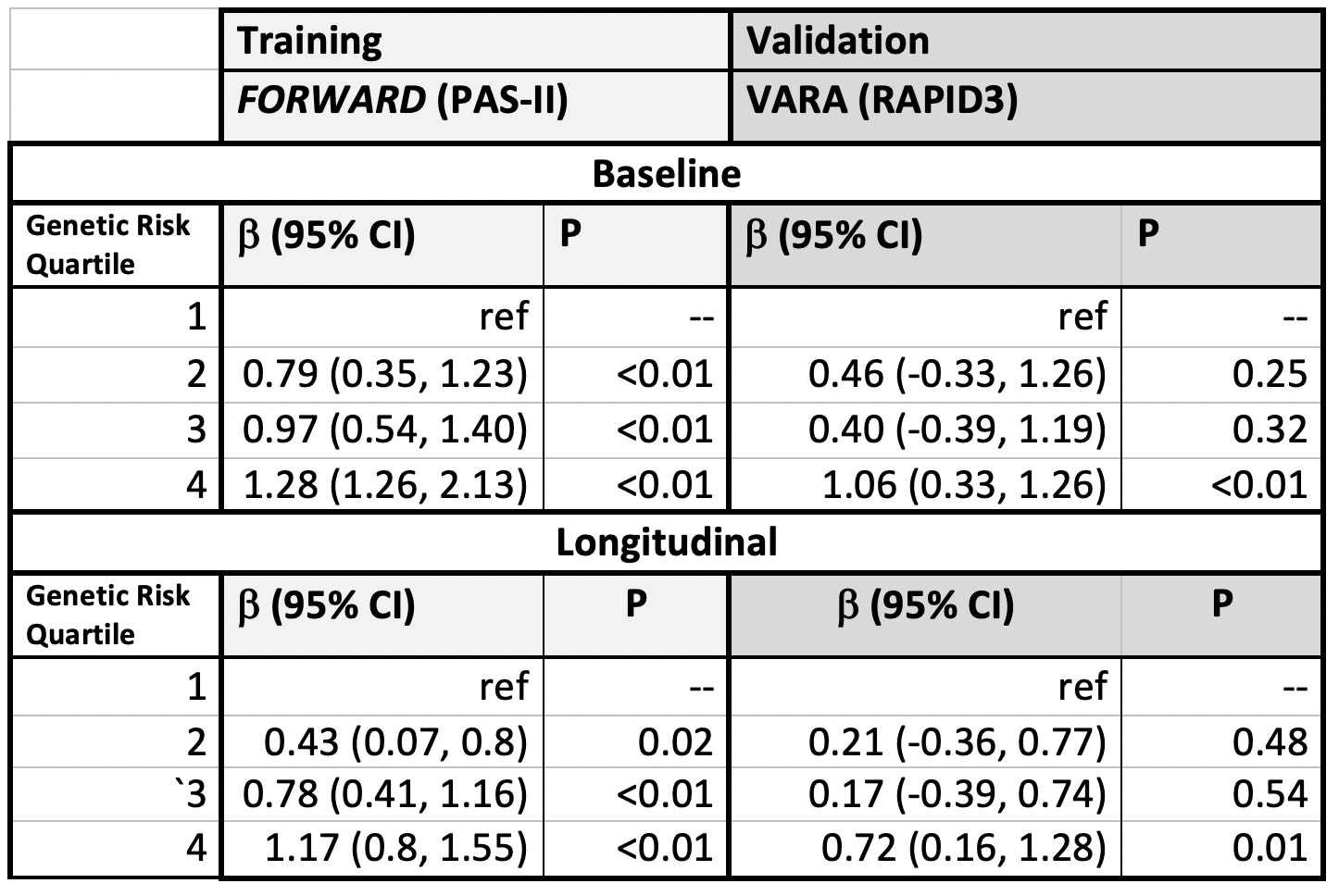
Results: This study included 765 patients from FORWARD (mean age 56.8 years, 89.4% female) and 2176 from VARA (mean age 71.7 years, 11.0% female). Several SNPs were associated with pain scores in each cohort, but no single SNP was associated in both cohorts. For example, VARA participants with the homozygous major allele of the BDNF rs6265 polymorphism (brain-derived neurotrophic factor) had less pain [B: -2.16 (95% CI: -4.06, -0.25, p=0.02)] compared to those with the homozygous minor allele, but a similar association was not observed in FORWARD.FORWARD participants with a higher GRS had significantly more pain, with those in the highest quartile having higher baseline pain scores [+2.44 (95% CI: (1.91, 2.97) p< 0.01 (range 0-10)] (Figure). VARA participants in the greatest quartile of GRS also had modestly higher baseline pain scores [+0.54 (95% CI: 0.20, 0.89), p< 0.01]. Participants in the highest GRS quartile from both cohorts also had significantly greater pain scores throughout follow-up. The GRS was associated with higher baseline and longitudinal patient-reported disease activity as measured by the PAS-II (FORWARD) and RAPID3 (VARA) (Table).
Conclusion: A genetic risk score based on previously identified pain-related SNPs modestly predicted greater pain in an external independent cohort. These observations suggest that more comprehensive genetic scores may eventually provide meaningful clinical value in understanding pain in RA patients. More accurate prediction may be possible through more advanced methods such as machine learning and with an increasing understanding of the genes involved in pain processing.
.jpg)
K. McMenamin: None; K. Wipfler: None; A. Wheeler: None; G. Cannon: None; K. Wysham: None; B. Sauer: None; B. England: Boehringer-Ingelheim, 2, 5; K. Michaud: None; T. Mikuls: Elsevier, 9, Horizon Therapeutics, 2, 5, Pfizer, 2, Sanofi, 2, UCB Pharma, 2, Wolters Kluwer Health (UpToDate), 9; J. Baker: CorEvitas, 2, Cumberland Pharma, 2, Horizon Pharmaceuticals, 5.
Background/Purpose: Pain in rheumatoid arthritis (RA) is multifactorial and involves processes beyond inflammation such as peripheral and central pain processing. Several genes have been associated with pain processing through studies of fibromyalgia (FM), but it is largely unknown how genetics may affect pain in other rheumatic diseases. The aim of this study was to determine if single nucleotide polymorphisms (SNPs) associated with FM also have an impact on pain and disease activity in patients with RA.
Methods: Participants with RA with whole genome SNP data were included from two independent cohorts: 1) FORWARD (National Data Bank for Rheumatic Diseases) and 2) Veterans Affairs RA registry (VARA). Linear regression was used to determine the relationship between 30 individual SNPs previously associated with FM (Jannsen et al. An Acad Bras Ciênc. 2021) and baseline pain scores in FORWARD adjusting for age, sex and race. A genetic risk score (GRS) was generated using all 30 SNPs, weighted by the coefficients for each SNP from this regression. Linear regression was then used to assess the relationship between the GRS and cross-sectional (baseline) pain (visual analogue scale, range 0-10) in FORWARD (training dataset) and VARA (validation dataset). Similarly, the association of GRS with pain during longitudinal follow-up was determined using regression with generalized estimating equations adjusting for age, sex and race. In addition, the associations between GRS and disease activity, at enrollment and during longitudinal follow-up in each study were determined.
Results: This study included 765 patients from FORWARD (mean age 56.8 years, 89.4% female) and 2176 from VARA (mean age 71.7 years, 11.0% female). Several SNPs were associated with pain scores in each cohort, but no single SNP was associated in both cohorts. For example, VARA participants with the homozygous major allele of the BDNF rs6265 polymorphism (brain-derived neurotrophic factor) had less pain [B: -2.16 (95% CI: -4.06, -0.25, p=0.02)] compared to those with the homozygous minor allele, but a similar association was not observed in FORWARD.FORWARD participants with a higher GRS had significantly more pain, with those in the highest quartile having higher baseline pain scores [+2.44 (95% CI: (1.91, 2.97) p< 0.01 (range 0-10)] (Figure). VARA participants in the greatest quartile of GRS also had modestly higher baseline pain scores [+0.54 (95% CI: 0.20, 0.89), p< 0.01]. Participants in the highest GRS quartile from both cohorts also had significantly greater pain scores throughout follow-up. The GRS was associated with higher baseline and longitudinal patient-reported disease activity as measured by the PAS-II (FORWARD) and RAPID3 (VARA) (Table).
Conclusion: A genetic risk score based on previously identified pain-related SNPs modestly predicted greater pain in an external independent cohort. These observations suggest that more comprehensive genetic scores may eventually provide meaningful clinical value in understanding pain in RA patients. More accurate prediction may be possible through more advanced methods such as machine learning and with an increasing understanding of the genes involved in pain processing.
.jpg)
Difference in baseline mean VAS pain score by quartile of genetic risk score for FORWARD (training) and VARA (validation) cohorts

Genetic risk score quartile and disease activity score at baseline and throughout all observations for FORWARD (training) and VARA (validation) datasets
K. McMenamin: None; K. Wipfler: None; A. Wheeler: None; G. Cannon: None; K. Wysham: None; B. Sauer: None; B. England: Boehringer-Ingelheim, 2, 5; K. Michaud: None; T. Mikuls: Elsevier, 9, Horizon Therapeutics, 2, 5, Pfizer, 2, Sanofi, 2, UCB Pharma, 2, Wolters Kluwer Health (UpToDate), 9; J. Baker: CorEvitas, 2, Cumberland Pharma, 2, Horizon Pharmaceuticals, 5.



