Poster Session B
Systemic lupus erythematosus (SLE)
Session: (0899–0933) SLE – Etiology & Pathogenesis Poster
0915: IL-16+ Is Abundantly Expressed by Kidney-infiltrating Myeloid and Lymphoid Cells in Lupus Nephritis: A Spatially Resolved Multiplexed Approach
Monday, November 13, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- AC
Alessandra Ida Celia, MD (she/her/hers)
Sapienza University of Rome
Rome, Rome, ItalyDisclosure(s): No financial relationships with ineligible companies to disclose
Abstract Poster Presenter(s)
Alessandra Ida Celia1, Xiaoping Yang2, Hana Minsky2, Silvia Malvica2, Michelle Petri3, the Accelerating Medicines Partnership in RA/SLE4, Avi Rosenberg5 and Andrea Fava2, 1John Hopkins University of Medicine, Rome, Italy, 2Johns Hopkins University, Baltimore, MD, 3Department of Medicine, Division of Rheumatology, Johns Hopkins University School of Medicine, Timonium, MD, 4Multiple, Multiple, 5Department of Pathology, The Johns Hopkins University School of Medicine, Baltimore, MD
Background/Purpose: We previously discovered that IL-16 is the urinary protein most strongly correlated with histological activity in lupus nephritis (LN), followed by neutrophil degranulation products such as PR3. IL-16 is a proinflammatory chemokine that binds to CD4 and CD9 which are also expressedby myeloid cells. IL-16 is a key mediator in renal ischemia-reperfusion injury. Furthermore, IL-16 polymorphisms carry a higher risk of SLE (OR 3.3-10.4), suggesting a potential causal role. Our hypothesis is that IL-16 critically amplifies the immune response in proliferative LN, thereby attracting myeloid populations that can ultimately lead to kidney damage. The aims of our study are to investigate IL-16+ cells in LN kidney, define their immunophenotype, and to explore the presence of PR3+ cells in the same spatial area in order to gain insight on the spatial relation between the two populations.
Methods: We performed 20-plex serial immunohistochemistry (sIHC) on one archival LN kidney biopsy (ISN/RPS class IV) to identify IL-16+ and PR3+ cells and evaluate the expression of multiple cell lineage markers. We utilized Indica HALO to perform image analysis, including deconvolution, cell segmentation, glomerular annotation, and quantitative histology.
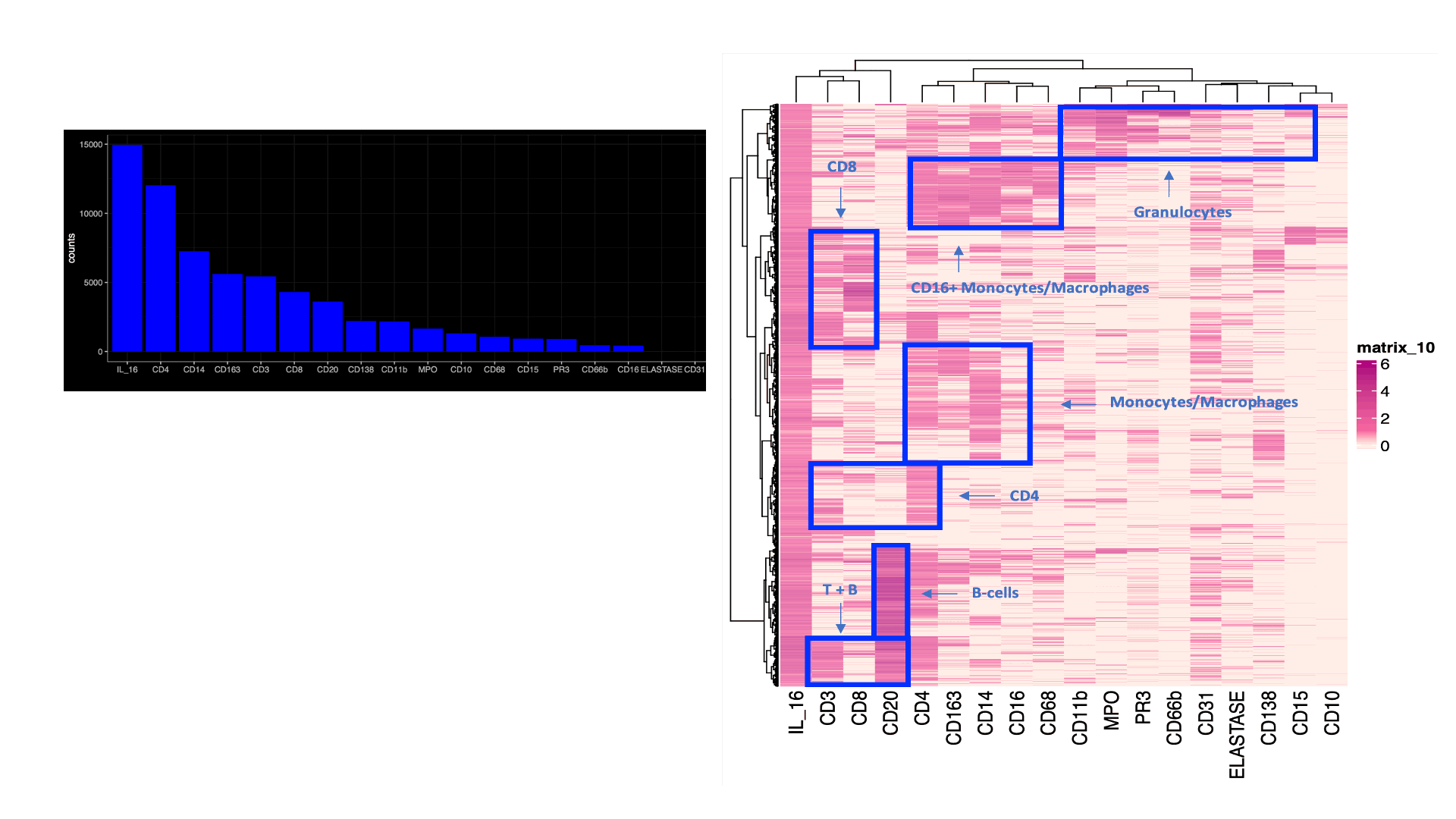
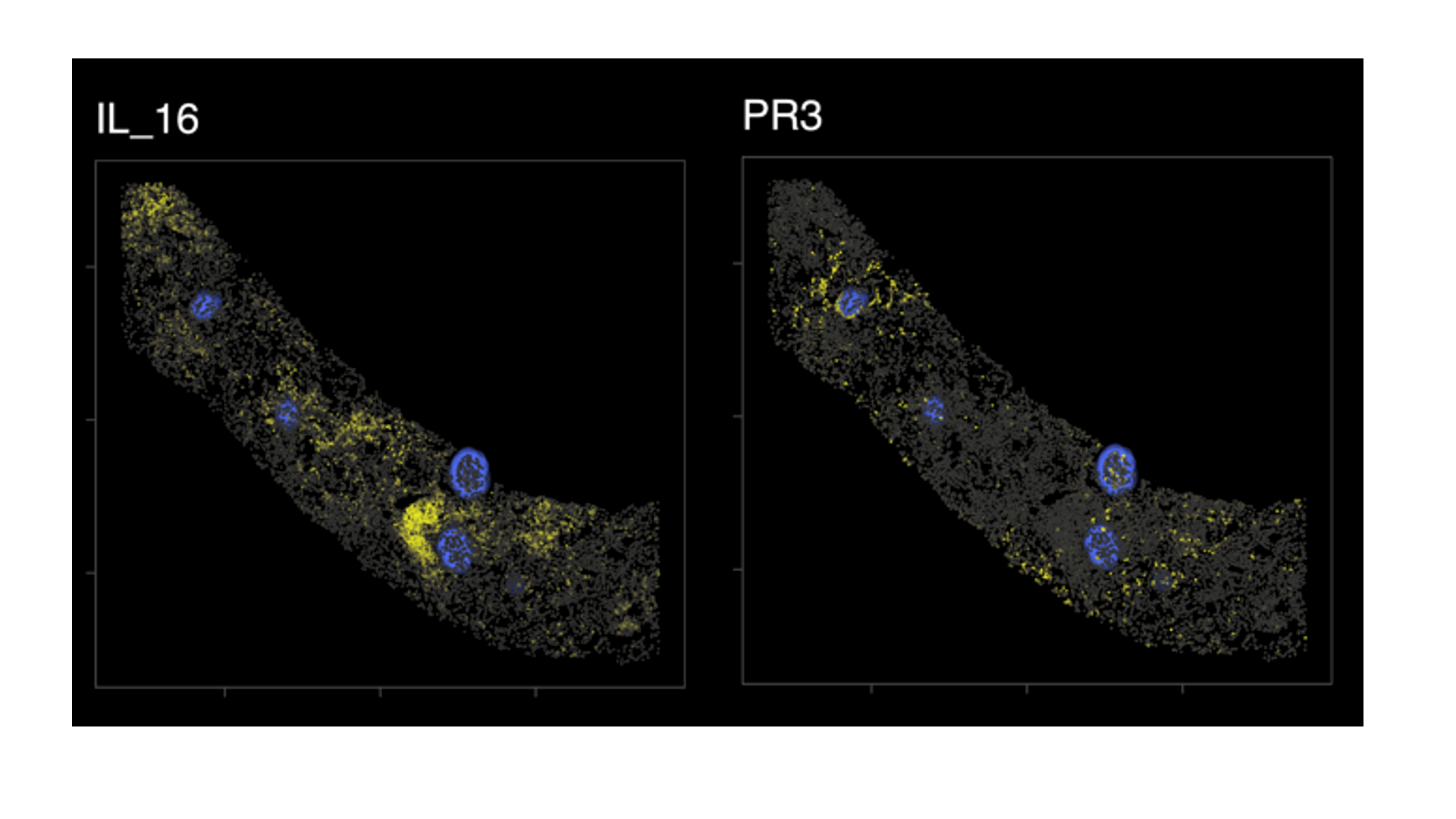
Results: A total of 95,619 cell objects present in the sample were analyzed. We observed an abundant IL-16+ infiltrate, mostly in the tubulointerstitium. Multiple kidney-infiltrating immune cell types produced IL-16. The co-expression of lineage markers by IL-16+ cells is summarized by Figure1A. Figure 1Bdisplays the heterogeneity of marker co-expression at the single cell level for IL-16+ cells. Most IL-16+ cells also expressed CD4, a ligand for IL-16. Figure 2 illustrates the tissue distribution of IL-16 and PR3+ cells showing how IL-16 and PR3 are not spatially related.
Conclusion: IL-16+ cells abundantly infiltrate the kidney in LN. IL-16 is producedby most types of immune cells including lymphoid and myeloid cells. There was no spatial relationship between IL-16+ and PR3+ cells suggesting no obvious chemotactic activity. The release of IL-16 by multiple immune cell types may promote and amplify LN.
A. Celia: None; X. Yang: None; H. Minsky: None; S. Malvica: None; M. Petri: Alexion, 1, Amgen, 1, AnaptysBio, 1, Annexon Bio, 1, Argenx, 1, Arhros-Focus Med/Ed, 6, AstraZeneca, 1, 5, Aurinia, 1, 5, 6, Axdev, 1, Biogen, 1, Boxer Capital, 2, Cabaletto Bio, 2, Caribou Biosciences, 2, CVS Health, 1, Eli Lilly, 1, 5, Emergent Biosolutions, 1, Exagen, 5, Exo Therapeutics, 2, Gilead Biosciences, 2, GlaxoSmithKlein(GSK), 1, 5, 6, Horizon Therapeutics, 2, Idorsia Pharmaceuticals, 2, IQVIA, 1, Janssen, 1, 5, Kira Pharmaceuticals, 2, MedShr, 6, Merck/EMD Serono, 1, Momenta Pharmaceuticals, 2, Nexstone Immunology, 2, Nimbus Lakshmi, 2, Proviant, 2, Sanofi, 2, Sinomab Biosciences, 2, Thermofisher, 5, UCB, 2; t. Accelerating Medicines Partnership in RA/SLE: None; A. Rosenberg: None; A. Fava: Annexon Biosciences, 2, Sanofi, 1.
Background/Purpose: We previously discovered that IL-16 is the urinary protein most strongly correlated with histological activity in lupus nephritis (LN), followed by neutrophil degranulation products such as PR3. IL-16 is a proinflammatory chemokine that binds to CD4 and CD9 which are also expressedby myeloid cells. IL-16 is a key mediator in renal ischemia-reperfusion injury. Furthermore, IL-16 polymorphisms carry a higher risk of SLE (OR 3.3-10.4), suggesting a potential causal role. Our hypothesis is that IL-16 critically amplifies the immune response in proliferative LN, thereby attracting myeloid populations that can ultimately lead to kidney damage. The aims of our study are to investigate IL-16+ cells in LN kidney, define their immunophenotype, and to explore the presence of PR3+ cells in the same spatial area in order to gain insight on the spatial relation between the two populations.
Methods: We performed 20-plex serial immunohistochemistry (sIHC) on one archival LN kidney biopsy (ISN/RPS class IV) to identify IL-16+ and PR3+ cells and evaluate the expression of multiple cell lineage markers. We utilized Indica HALO to perform image analysis, including deconvolution, cell segmentation, glomerular annotation, and quantitative histology.
Results: A total of 95,619 cell objects present in the sample were analyzed. We observed an abundant IL-16+ infiltrate, mostly in the tubulointerstitium. Multiple kidney-infiltrating immune cell types produced IL-16. The co-expression of lineage markers by IL-16+ cells is summarized by Figure1A. Figure 1Bdisplays the heterogeneity of marker co-expression at the single cell level for IL-16+ cells. Most IL-16+ cells also expressed CD4, a ligand for IL-16. Figure 2 illustrates the tissue distribution of IL-16 and PR3+ cells showing how IL-16 and PR3 are not spatially related.
Conclusion: IL-16+ cells abundantly infiltrate the kidney in LN. IL-16 is producedby most types of immune cells including lymphoid and myeloid cells. There was no spatial relationship between IL-16+ and PR3+ cells suggesting no obvious chemotactic activity. The release of IL-16 by multiple immune cell types may promote and amplify LN.

Figure1. IL-16+ cells immune phenotype
(A) Coexpression of lineage markers in IL-16+ cells. Values indicate number (count) of positive cells. (B) Heatmaps displaying the heterogeneity of marker coexpression at the single cell level for IL-16+ cells (B). Data from one representative kidney biopsy (class IV lupus nephritis, NIH Activity Index 8/24, Chronicity Index 6/12).
(A) Coexpression of lineage markers in IL-16+ cells. Values indicate number (count) of positive cells. (B) Heatmaps displaying the heterogeneity of marker coexpression at the single cell level for IL-16+ cells (B). Data from one representative kidney biopsy (class IV lupus nephritis, NIH Activity Index 8/24, Chronicity Index 6/12).

Figure2. Spatial distribution of IL-16+ and PR3+ cells
Biopsy detail mapping IL-16 and PR3+ cells (in yellow); glomeruli in blue. There was no obvious spatial relation between IL16+ and PR3+ cells. Data from one representative kidney biopsy (class IV lupus nephritis, NIH Activity Index 8/24, Chronicity Index 6/12).
Biopsy detail mapping IL-16 and PR3+ cells (in yellow); glomeruli in blue. There was no obvious spatial relation between IL16+ and PR3+ cells. Data from one representative kidney biopsy (class IV lupus nephritis, NIH Activity Index 8/24, Chronicity Index 6/12).
A. Celia: None; X. Yang: None; H. Minsky: None; S. Malvica: None; M. Petri: Alexion, 1, Amgen, 1, AnaptysBio, 1, Annexon Bio, 1, Argenx, 1, Arhros-Focus Med/Ed, 6, AstraZeneca, 1, 5, Aurinia, 1, 5, 6, Axdev, 1, Biogen, 1, Boxer Capital, 2, Cabaletto Bio, 2, Caribou Biosciences, 2, CVS Health, 1, Eli Lilly, 1, 5, Emergent Biosolutions, 1, Exagen, 5, Exo Therapeutics, 2, Gilead Biosciences, 2, GlaxoSmithKlein(GSK), 1, 5, 6, Horizon Therapeutics, 2, Idorsia Pharmaceuticals, 2, IQVIA, 1, Janssen, 1, 5, Kira Pharmaceuticals, 2, MedShr, 6, Merck/EMD Serono, 1, Momenta Pharmaceuticals, 2, Nexstone Immunology, 2, Nimbus Lakshmi, 2, Proviant, 2, Sanofi, 2, Sinomab Biosciences, 2, Thermofisher, 5, UCB, 2; t. Accelerating Medicines Partnership in RA/SLE: None; A. Rosenberg: None; A. Fava: Annexon Biosciences, 2, Sanofi, 1.



