Poster Session A
Immunobiology
Session: (0040–0064) Innate Immunity Poster
0051: CD14+CD64+ Classical Monocytes Are the Main Producers of IL-23 at the Enthesis
Sunday, November 12, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- NM
Nicole McDermott, PhD, BSc
University of Leeds
Leeds, United KingdomDisclosure information not submitted.
Abstract Poster Presenter(s)
Nicole McDermott1, Thomas Macleod1, Ala Altaie1, Liz Straszynski2, Robert Dunsmuir3, Vishal Borse3, Peter Loughenbury3, Davide Simone4, Stevephen Sansom4, Christopher Buckley4 and Dennis McGonagle5, 1University of Leeds, Leeds Institute of Rheumatic and Musculoskeletal Medicine, Leeds, United Kingdom, 2Leeds Institute of Medical Research at St. James’s, Leeds, United Kingdom, 3Leeds Teaching Hospital NHS Trust, Leeds, United Kingdom, 4Kennedy Institute of Rheumatology, Oxford, United Kingdom, 5Leeds Biomedical Research Centre, University of Leeds, Leeds, United Kingdom
Background/Purpose: IL-23 is a key cytokine involved in diseases such as psoriasis, psoriatic arthritis and spondyloarthropathies (SpA) and inflammatory bowel disease.IL-23 is produced by activated monocytes, dendritic cells and neutrophils. Guselkumab, IL-23 p19 subunit inhibitor binds to native FcR1 (CD64) but Risankizumab, another p19 blocker, has a mutated Fc domain and lacks this binding. Entheses are relatively avascular sites and may be difficult for antibody penetration and normal entheseal macrophages have inducible IL-23 protein expression. In this work we investigated the phenotype and functionality of normal and activated entheseal macrophages to explore CD64 expression and which monocyte population had predominant IL-23 expression.
Methods: Immune cell populations were flushed from spinous process; samples were obtained from patients undergoing surgery for spinal decompression with no known chronic inflammation. Cells are sorted into two populations CD64+ and CD64-, sorting was gated on live cell and CD3+ depleted. The populations that were sorted are CD64+CD14+CD16- and CD64-CD14-CD16+. Cells were cultured for 48hrs with LPS (100ng/ml) & IFN-γ (50ng/ml) and then assessed for IL-23 production by ELISA and cells phenotyped to assess their CD64 profile (N=4). Single cell sequencing has been performed on bone and soft tissue from the enthesis following collagenase digest and CD64 expression identified (N=4).
Results: Sorting profiles indicated that the CD64+ cells were CD14+CD16- and CD64- cells were CD14-CD16+, the enthesis showed some cells that are CD64+ have intermediate CD14 expression (Figure 1). Following 48hrs in culture cells maintained their respective CD64 expression profile confirming that CD64 expression does not fluctuate upon cell activation (Figure 2). Analysis of the supernatant reveals that IL-23 expression distinctly comes from the CD64+CD14+ cells from the enthesis and predominantly from the PBMCs following activation with LPS & IFN-γ (Figure 2). Single cell analysis confirms that CD64 expression is found on the conventional and inflammatory monocytes which makes up approximately 20% of the cell population and even in health a proportion of these cells had IL23A gene expression which encodes the P19 subunit (Figure 3).
Conclusion: In this work, we have shown that CD64+CD14+ conventional monocytes are the main producers of IL-23 at the axial enthesis. This has been confirmed by flow cytometry and single cell analysis showing that these make up approximately 20% of the total cell population. Given that the key entheseal cell producing IL-23 also expresses CD64, the receptor for the rapid removal of Guselkumab bound IL-23 at the enthesis. These findings highlight that there could be differences between Guselkumab and Risankizumab in IL-23 neutralisation at the key enthesis territory.
N. McDermott: None; T. Macleod: None; A. Altaie: None; L. Straszynski: None; R. Dunsmuir: None; V. Borse: None; P. Loughenbury: None; D. Simone: None; S. Sansom: None; C. Buckley: None; D. McGonagle: AbbVie, 2, 5, 6, Celgene, 2, 5, 6, Janssen, 2, 5, 6, Merck, 2, 5, 6, Novartis, 2, 5, 6, Pfizer, 2, 5, 6, UCB, 2, 5, 6.
Background/Purpose: IL-23 is a key cytokine involved in diseases such as psoriasis, psoriatic arthritis and spondyloarthropathies (SpA) and inflammatory bowel disease.IL-23 is produced by activated monocytes, dendritic cells and neutrophils. Guselkumab, IL-23 p19 subunit inhibitor binds to native FcR1 (CD64) but Risankizumab, another p19 blocker, has a mutated Fc domain and lacks this binding. Entheses are relatively avascular sites and may be difficult for antibody penetration and normal entheseal macrophages have inducible IL-23 protein expression. In this work we investigated the phenotype and functionality of normal and activated entheseal macrophages to explore CD64 expression and which monocyte population had predominant IL-23 expression.
Methods: Immune cell populations were flushed from spinous process; samples were obtained from patients undergoing surgery for spinal decompression with no known chronic inflammation. Cells are sorted into two populations CD64+ and CD64-, sorting was gated on live cell and CD3+ depleted. The populations that were sorted are CD64+CD14+CD16- and CD64-CD14-CD16+. Cells were cultured for 48hrs with LPS (100ng/ml) & IFN-γ (50ng/ml) and then assessed for IL-23 production by ELISA and cells phenotyped to assess their CD64 profile (N=4). Single cell sequencing has been performed on bone and soft tissue from the enthesis following collagenase digest and CD64 expression identified (N=4).
Results: Sorting profiles indicated that the CD64+ cells were CD14+CD16- and CD64- cells were CD14-CD16+, the enthesis showed some cells that are CD64+ have intermediate CD14 expression (Figure 1). Following 48hrs in culture cells maintained their respective CD64 expression profile confirming that CD64 expression does not fluctuate upon cell activation (Figure 2). Analysis of the supernatant reveals that IL-23 expression distinctly comes from the CD64+CD14+ cells from the enthesis and predominantly from the PBMCs following activation with LPS & IFN-γ (Figure 2). Single cell analysis confirms that CD64 expression is found on the conventional and inflammatory monocytes which makes up approximately 20% of the cell population and even in health a proportion of these cells had IL23A gene expression which encodes the P19 subunit (Figure 3).
Conclusion: In this work, we have shown that CD64+CD14+ conventional monocytes are the main producers of IL-23 at the axial enthesis. This has been confirmed by flow cytometry and single cell analysis showing that these make up approximately 20% of the total cell population. Given that the key entheseal cell producing IL-23 also expresses CD64, the receptor for the rapid removal of Guselkumab bound IL-23 at the enthesis. These findings highlight that there could be differences between Guselkumab and Risankizumab in IL-23 neutralisation at the key enthesis territory.

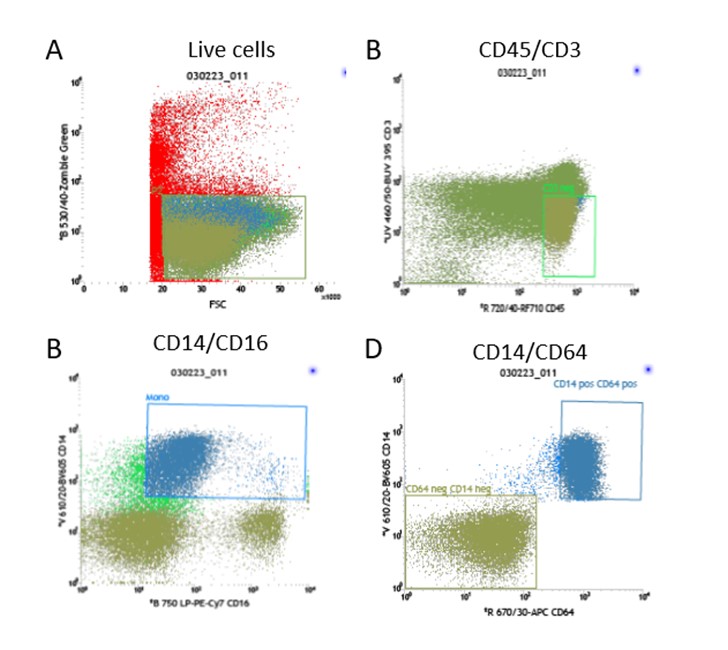
Figure 1: Gating strategy for sorting CD64+ and CD64- cells.
Cells flushed from the spinous process are stained for cell sorting using the panel of markers: Live/dead-Zombie Green, CD45-RF710, CD3-BUV395, CD14-BV605, CD16-PE-Cy7 & CD64-APC. The gating strategy uses FSC/SSC to remove the granulocytes and then gates on the live cells (A), from the live cell population the CD3+ cells are gated out (B) which we have shown previously to be CD64 negative. Cells are then separated by CD14 and CD16 (C), the CD64+ cells collected are CD14+ as shown in the bottom left plot and the CD64- cells are CD14- (D).
Cells flushed from the spinous process are stained for cell sorting using the panel of markers: Live/dead-Zombie Green, CD45-RF710, CD3-BUV395, CD14-BV605, CD16-PE-Cy7 & CD64-APC. The gating strategy uses FSC/SSC to remove the granulocytes and then gates on the live cells (A), from the live cell population the CD3+ cells are gated out (B) which we have shown previously to be CD64 negative. Cells are then separated by CD14 and CD16 (C), the CD64+ cells collected are CD14+ as shown in the bottom left plot and the CD64- cells are CD14- (D).

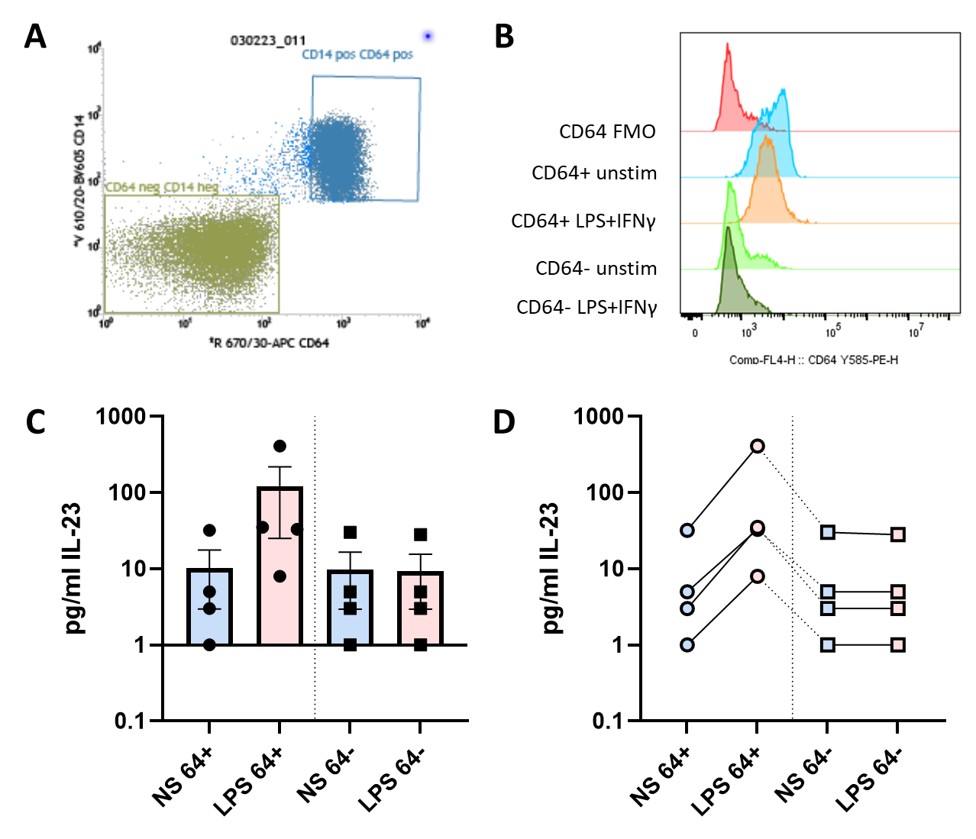
Figure 2: IL-23 production is from activated entheseal monocytes.
Sorting of entheseal CD64+ cells correlated with CD14 positivity and CD64- cells correlated with CD14 negativity (A), these two cell populations were put into culture for 48hrs stimulated with LPS (100ng/mL) and IFN-γ (50ng/mL) or with no stimulation. After 48hrs in culture CD64 profiles were reassessed by flow cytometry (B), from both CD64+ and CD64- populations, expression was compared to a CD64 FMO. Supernatants collected after 48hrs were assessed via ELISA to measure the levels of IL-23 N=4 (C), each of the four donors was plotted and shows levels of IL-23 for each condition follows the same trend (D).
Sorting of entheseal CD64+ cells correlated with CD14 positivity and CD64- cells correlated with CD14 negativity (A), these two cell populations were put into culture for 48hrs stimulated with LPS (100ng/mL) and IFN-γ (50ng/mL) or with no stimulation. After 48hrs in culture CD64 profiles were reassessed by flow cytometry (B), from both CD64+ and CD64- populations, expression was compared to a CD64 FMO. Supernatants collected after 48hrs were assessed via ELISA to measure the levels of IL-23 N=4 (C), each of the four donors was plotted and shows levels of IL-23 for each condition follows the same trend (D).

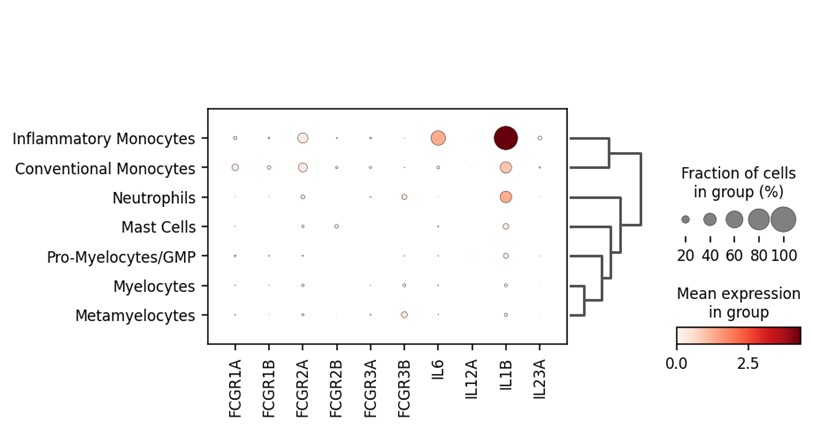
Figure 3: Singe-cell sequencing reveals gene expression for FCGR1A/B and IL23A is only present in the monocyte populations.
Single-cell sequencing was performed on spinal entheseal bone and soft tissue, samples were prepared by performing a collagenase digest. Bone and soft tissue were analysed, and cluster analysis reveals that the FGR1A and FCGR2A genes which encode for CD64 are present in approximately 20% of the conventional and inflammatory monocytes, this also correlates with expression of the IL23A gene encoding for IL-23 N=4. Together this data confirms what has been seen at the protein level.
Single-cell sequencing was performed on spinal entheseal bone and soft tissue, samples were prepared by performing a collagenase digest. Bone and soft tissue were analysed, and cluster analysis reveals that the FGR1A and FCGR2A genes which encode for CD64 are present in approximately 20% of the conventional and inflammatory monocytes, this also correlates with expression of the IL23A gene encoding for IL-23 N=4. Together this data confirms what has been seen at the protein level.
N. McDermott: None; T. Macleod: None; A. Altaie: None; L. Straszynski: None; R. Dunsmuir: None; V. Borse: None; P. Loughenbury: None; D. Simone: None; S. Sansom: None; C. Buckley: None; D. McGonagle: AbbVie, 2, 5, 6, Celgene, 2, 5, 6, Janssen, 2, 5, 6, Merck, 2, 5, 6, Novartis, 2, 5, 6, Pfizer, 2, 5, 6, UCB, 2, 5, 6.



