Poster Session A
Periodic fever syndromes, autoinflammatory diseases, Still’s disease and MAS/HLH
Session: (0252–0282) Miscellaneous Rheumatic & Inflammatory Diseases Poster I
0282: DNA Methylation Markers in Peripheral Blood as Effective Diagnostic Biomarkers for Adult-onset Still’s Disease
Sunday, November 12, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- SR
Shijia Rao
The Second Xiangya Hospital, Central South University
Changsha, ChinaDisclosure information not submitted.
Abstract Poster Presenter(s)
Shijia Rao1, Bo Zhang2, Yuwei Li3, Tian Zhou1, Xiaoli Shi1, Wei Shi4, Yongfang Jiang1, Wenlong Wang1, Yun Xu1, Yi Tian1, Min Zhang1, Jing Ma1, Hongyu Luo1, Hui Luo4, Hongjun Zhao4, Fen Li1, Guanghui Ling1, Jing Tian1, Xiaojing Kang5, Junqin Liang5, Aijun Chen6, Shuang Chen6, Jianjun Qiao7, Gang Wang8, Lei wang8, Bing Li8, Youkun Lin9, Sijian Wen9, Zhiqiang Song10, Min Zhang10, Jun Liu4, Yixin Tan1, Haijing Wu1, Hai Long1, Ming Zhao11 and Qianjin Lu12, 1The Second Xiangya Hospital, Central South University, Changsha, China, 2Chinese Academy of Medical Sciences & Peking Union Medical College, Nanjing, China, 3University of Science and Technology of China, Hefei, China, 4Xiangya Hospital, Central South University, Changsha, China, 5People's Hospital of Xinjiang Uygur Autonomous Region, Urumqi, China, 6The First Affiliated Hospital of Chongqing Medical University, Chongqing, China, 7The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China, 8Xijing Hospital, Fourth Military Medical University, Xi'an, China, 9The First Affiliated Hospital of Guangxi Medical University, Nanning, China, 10Southwest Hospital, Third Military Medical University (Army Medical University), Chongqing, China, 11Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, China, 12Hospital for Skin Diseases, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Changsha, China
Background/Purpose: Adult-onset still's disease (AOSD) is an auto-inflammatory disease affecting multiple systems. The diagnostic procedures of AOSD are still a challenge. We aimed to find an effective DNA methylation biomarker for AOSD diagnosis in this study
Methods: Differential methylation CpG sites (DMSs) were selected from DNA methylation array comprised of a discovery cohort including 50 AOSD patients, 49 healthy controls (HCs), 24 T cell lymphoma (TL) patients, 24 sepsis (SP) patients and 24 drug eruption (DE) patients. A cohort of 96 AOSD patients, 92 HCs, 50 rheumatoid arthritis (RA) patients, 28 SP patients, 21 DE patients and 62 TL patients was used for validation via pyrosequencing. Subsequently, four logistic regression prediction models were performed.
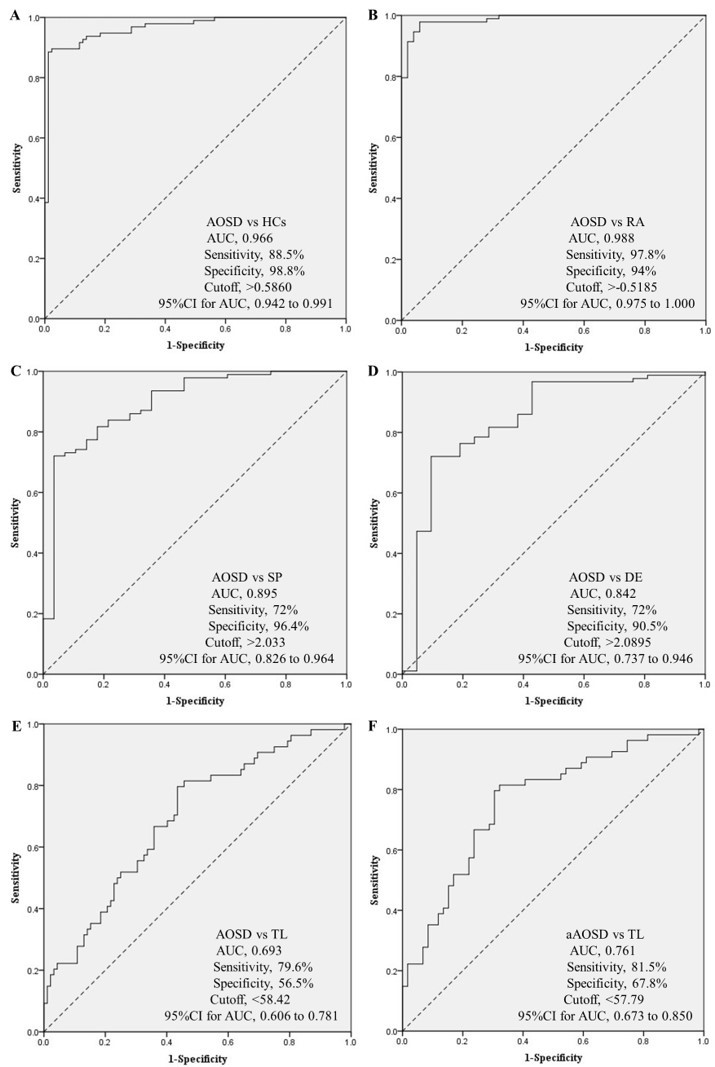
Results: Nine DMSs were selected from the discovery cohort via the DNA methylation array incorporating Least Absolute Shrinkage and Selector Operation (LASSO). We analyzed the methylation levels of these CpG sites by pyrosequencing in the validation cohort, and calculated four logistic regression prediction models in order to differentiate AOSD from the HCs (AUC: 0.964, sensitivity: 98.5%, specificity: 98.9%), the RA group (AUC: 0.988, sensitivity: 97.8%, specificity: 94%), the SP group (AUC: 0.885, sensitivity: 77.1%, specificity: 92.9%) and the DE group (AUC: 0.842, sensitivity: 72%, specificity: 90.5%) respectively. In addition, the methylation level of cg14887853 could be used to distinguish AOSD (AUC: 0.693, sensitivity: 79.6%, specificity: 59.5%) and active AOSD (AUC: 0.761, sensitivity: 81.5%, specificity: 67.8%) from TL respectively.
Conclusion: The DNA methylation levels of CpG sites can be used as biomarker for diagnosis of AOSD.
S. Rao: None; B. Zhang: None; Y. Li: None; T. Zhou: None; X. Shi: None; W. Shi: None; Y. Jiang: None; W. Wang: None; Y. Xu: None; Y. Tian: None; M. Zhang: None; J. Ma: None; H. Luo: None; H. Luo: None; H. Zhao: None; F. Li: None; G. Ling: None; J. Tian: None; X. Kang: None; J. Liang: None; A. Chen: None; S. Chen: None; J. Qiao: None; G. Wang: None; L. wang: None; B. Li: None; Y. Lin: None; S. Wen: None; Z. Song: None; M. Zhang: None; J. Liu: None; Y. Tan: None; H. Wu: None; H. Long: None; M. Zhao: None; Q. Lu: None.
Background/Purpose: Adult-onset still's disease (AOSD) is an auto-inflammatory disease affecting multiple systems. The diagnostic procedures of AOSD are still a challenge. We aimed to find an effective DNA methylation biomarker for AOSD diagnosis in this study
Methods: Differential methylation CpG sites (DMSs) were selected from DNA methylation array comprised of a discovery cohort including 50 AOSD patients, 49 healthy controls (HCs), 24 T cell lymphoma (TL) patients, 24 sepsis (SP) patients and 24 drug eruption (DE) patients. A cohort of 96 AOSD patients, 92 HCs, 50 rheumatoid arthritis (RA) patients, 28 SP patients, 21 DE patients and 62 TL patients was used for validation via pyrosequencing. Subsequently, four logistic regression prediction models were performed.
Results: Nine DMSs were selected from the discovery cohort via the DNA methylation array incorporating Least Absolute Shrinkage and Selector Operation (LASSO). We analyzed the methylation levels of these CpG sites by pyrosequencing in the validation cohort, and calculated four logistic regression prediction models in order to differentiate AOSD from the HCs (AUC: 0.964, sensitivity: 98.5%, specificity: 98.9%), the RA group (AUC: 0.988, sensitivity: 97.8%, specificity: 94%), the SP group (AUC: 0.885, sensitivity: 77.1%, specificity: 92.9%) and the DE group (AUC: 0.842, sensitivity: 72%, specificity: 90.5%) respectively. In addition, the methylation level of cg14887853 could be used to distinguish AOSD (AUC: 0.693, sensitivity: 79.6%, specificity: 59.5%) and active AOSD (AUC: 0.761, sensitivity: 81.5%, specificity: 67.8%) from TL respectively.
Conclusion: The DNA methylation levels of CpG sites can be used as biomarker for diagnosis of AOSD.

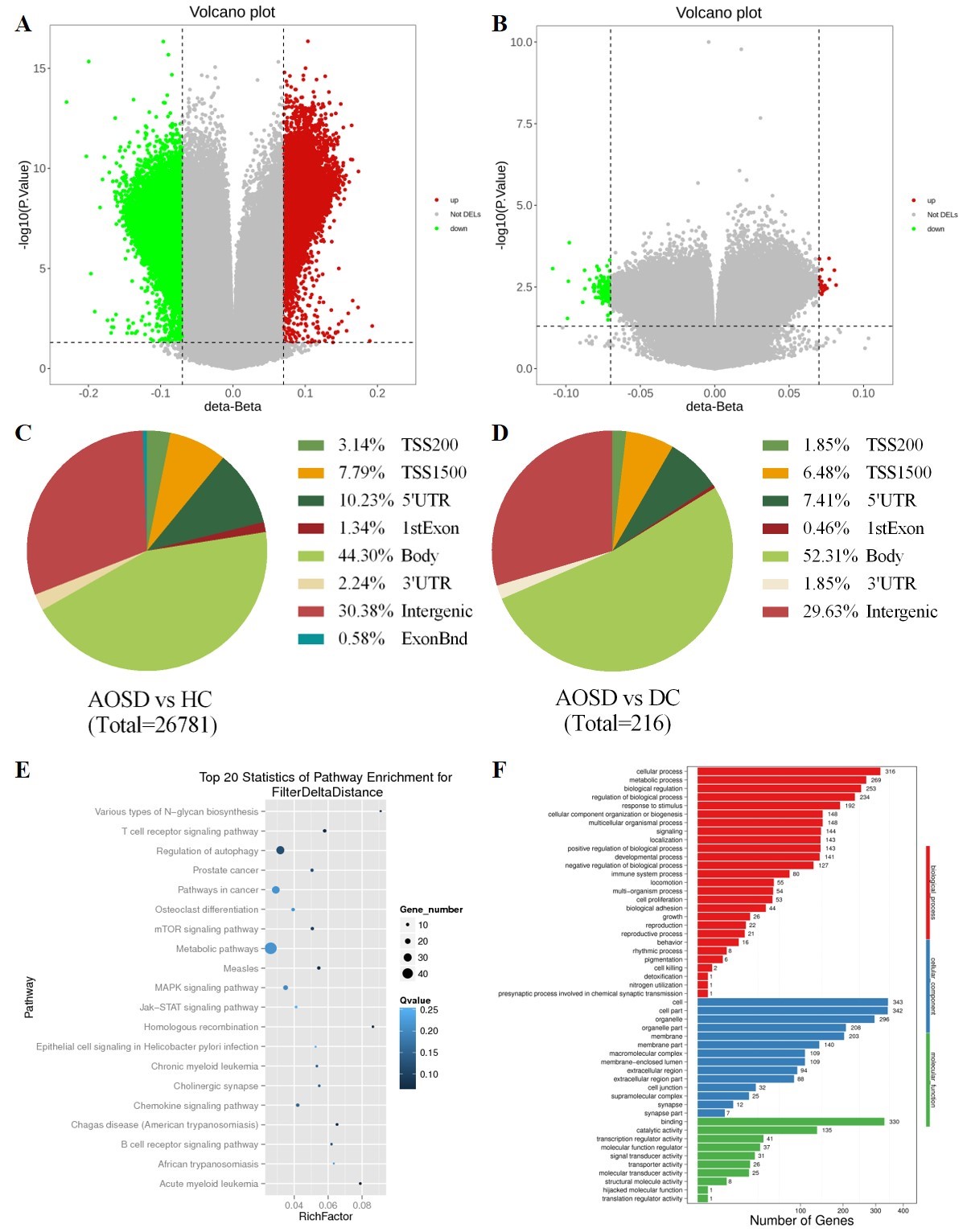
Volcano plots, location information, KEGG pathway analysis and GO analysis of differentially methylated CpG sites (DMSs) in AOSD. A Volcano plot of the distribution of DMSs between AOSD and HC group. B Volcano plot of the distribution of DMSs between AOSD and DC groups. C Location information of DMSs on genome E Top 20 statistic of pathway enrichment of DMSs between AOSD and HC group. F GO analysis DMSs between AOSD and HC group.

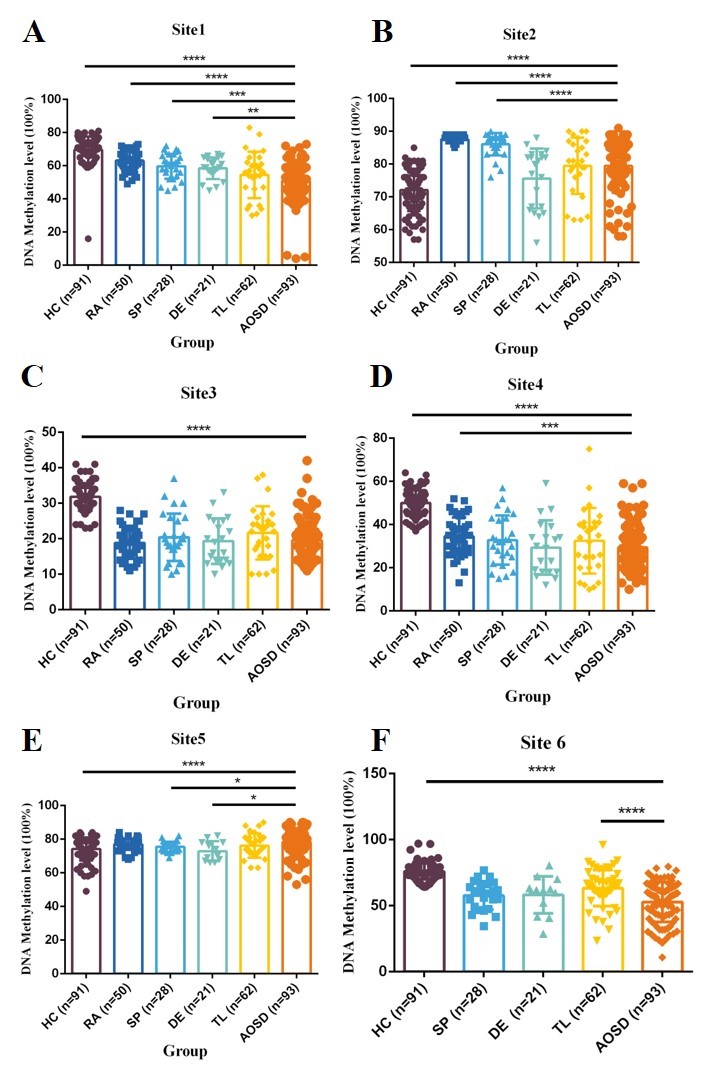
DNA methylation level of the identified six differential methylated sites (DMSs) between AOSD and healthy controls (HCs)/ disease controls (DCs) in validation cohort. A It shows that methylation level of Site1 is significantly lower in the blood of AOSD patients than that in HCs, patients with rheumatoid arthritis (RA), patients with sepsis (SP) and patients with drug eruption (DE). B It shows that methylation level of Site2 is significantly lower in the blood of AOSD patients than that in patients with RA and patients with SP, and is significantly higher in the blood of AOSD patients than that in HCs. C It shows that methylation level of Site3 is significantly lower in the blood of AOSD patients than that in HCs. D It shows that methylation level of Site4 is significantly lower in the blood of AOSD patients than that in HCs and patients with RA. E It shows that methylation level of Site5 is significantly higher in the blood of AOSD patients than that in HCs, patients with SP and patients with drug eruption (DE). F It shows that methylation level of Site6 is significantly lower in the blood of AOSD patients than that in HCs and patients with T cell lymphoma (TL).

Receiver operating characteristic (ROC) curves analysis of logistic regression prediction model. A show the ROC curve of the logistic regression prediction model based on Site1, Site3, Site4 and Site5 in patients with AOSD compared with HCs. B show the ROC curve of the logistic regression prediction model based on Site1, Site2, Site3 and Site4 in patients with AOSD compared with patients with RA. C show the ROC curve of the logistic regression prediction model based on Site1, Site2, and Site3 in patients with AOSD compared with patients with SP. D show the ROC curve of the logistic regression prediction model based on Site1, Site3, Site4 and Site5 in patients with AOSD compared with patients with DE. E show the ROC curve of Site 6 in patients with AOSD compared with patients with TL. F show the ROC curve of Site 6 in AOSD patients in active phase compared with patients with TL.
S. Rao: None; B. Zhang: None; Y. Li: None; T. Zhou: None; X. Shi: None; W. Shi: None; Y. Jiang: None; W. Wang: None; Y. Xu: None; Y. Tian: None; M. Zhang: None; J. Ma: None; H. Luo: None; H. Luo: None; H. Zhao: None; F. Li: None; G. Ling: None; J. Tian: None; X. Kang: None; J. Liang: None; A. Chen: None; S. Chen: None; J. Qiao: None; G. Wang: None; L. wang: None; B. Li: None; Y. Lin: None; S. Wen: None; Z. Song: None; M. Zhang: None; J. Liu: None; Y. Tan: None; H. Wu: None; H. Long: None; M. Zhao: None; Q. Lu: None.



