Poster Session A
Genetics, genomics and proteomics
Session: (0013–0039.5) Genetics, Genomics & Proteomics Poster
0032: Quantification of the Escape from X Chromosome Inactivation with the Million Cell-scale Human Blood Single-cell RNA-seq Datasets Reveals Heterogeneity of Escape Across Immune Cells and Escape in the Autoimmune Disease Conditions
Sunday, November 12, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- YT
Yoshihiko Tomofuji, MD (he/him/his)
Osaka University
Osaka, JapanDisclosure information not submitted.
Abstract Poster Presenter(s)
Yoshihiko Tomofuji1, Ryuya Edahiro1, Yuya Shirai1, Kyuto Sonehara2, Qingbo Wang1, Atsushi Kumanogoh1 and Yukinori Okada3, 1Osaka University, Suita, Japan, 2University of Tokyo, Tokyo, Japan, 3University of Tokyo / Osaka University / RIKEN, Tokyo, Japan
Background/Purpose: Understanding the differences in the immune system between sexes is important to elucidate the pathogenesis of autoimmune diseases which are more prevalent in females than males. One of the two X chromosomes of females is silenced through X chromosome inactivation (XCI) to compensate for the difference in the X chromosome dosage between sexes. There exist several genes which 'escape' from XCI, which could contribute to the differential gene expression between sexes. The differences in the escape across cell types are still poorly characterized, especially across immune cell types, even though a large number of immune-related genes are located on the X chromosome. Although previous studies suggested that abnormal escape of a few immune-related genes could contribute to autoimmune diseases, systematic evaluation of the association between escape and autoimmune diseases has not been conducted.
Methods: We investigated the escape across immune cell types utilizing the largest scale scRNA-seq datasets
(1) ~1,000,000 peripheral blood mononuclear cells (PBMCs) from 489 East Asian participants (Asian Immune Diversity Network), (2) ~1,000,000 PBMCs from 147 Japanese (72 COVID19 patients and 75 healthy participants) (3) ~1,000,000 PBMCs from 162 systematic lupus erythematosus patients and 99 healthy participants including East Asian and European.
We performed differential expression gene (DEG) analysis between sexes across immune cell types. In addition, we newly developed a method, single-cell Level inactivated X chromosome mapping (scLinaX), which directly quantifies gene expression from the inactivated X chromosome (Xi). Although the previous studies required plate-based scRNA-seq data which had rich per-cell information at the cost of throughput and cost efficiency for analyzing escape, scLinaX could be applied to the droplet-based scRNA-seq data which are high throughput and widely used.
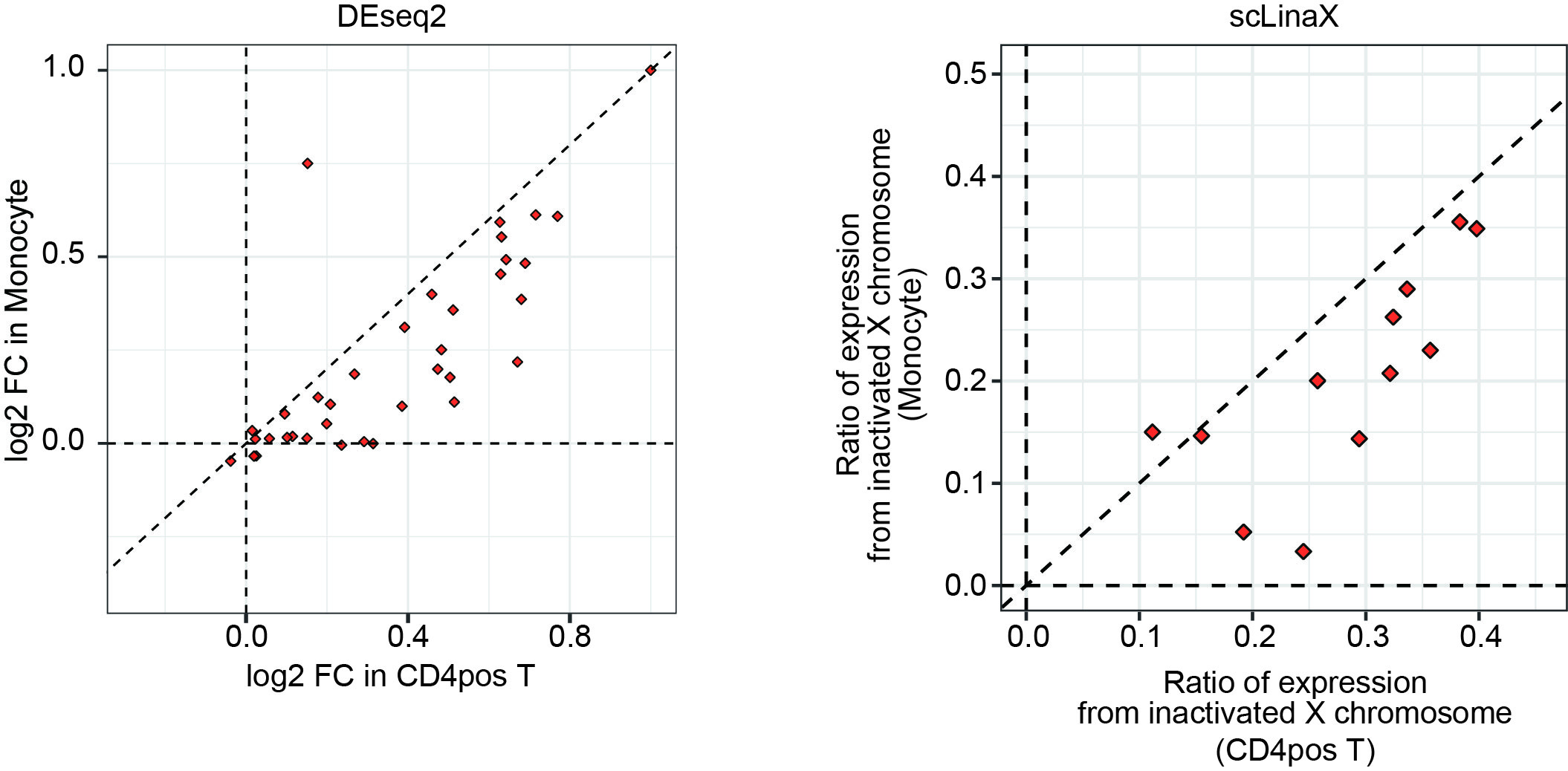
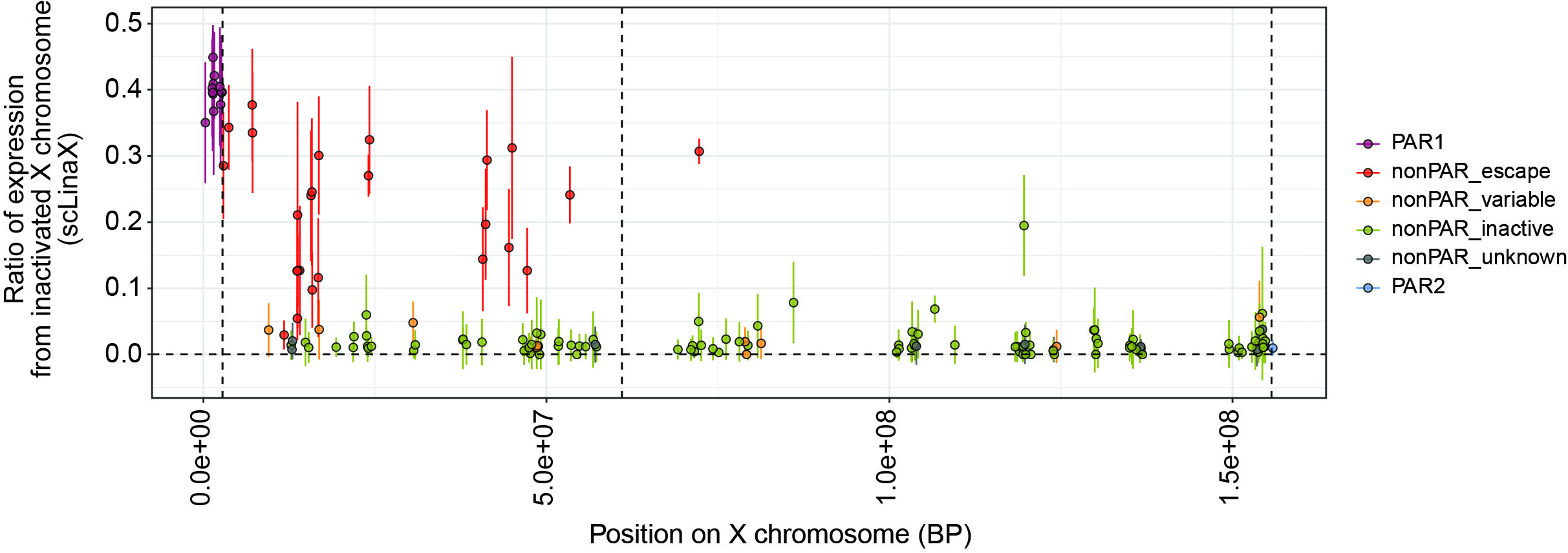
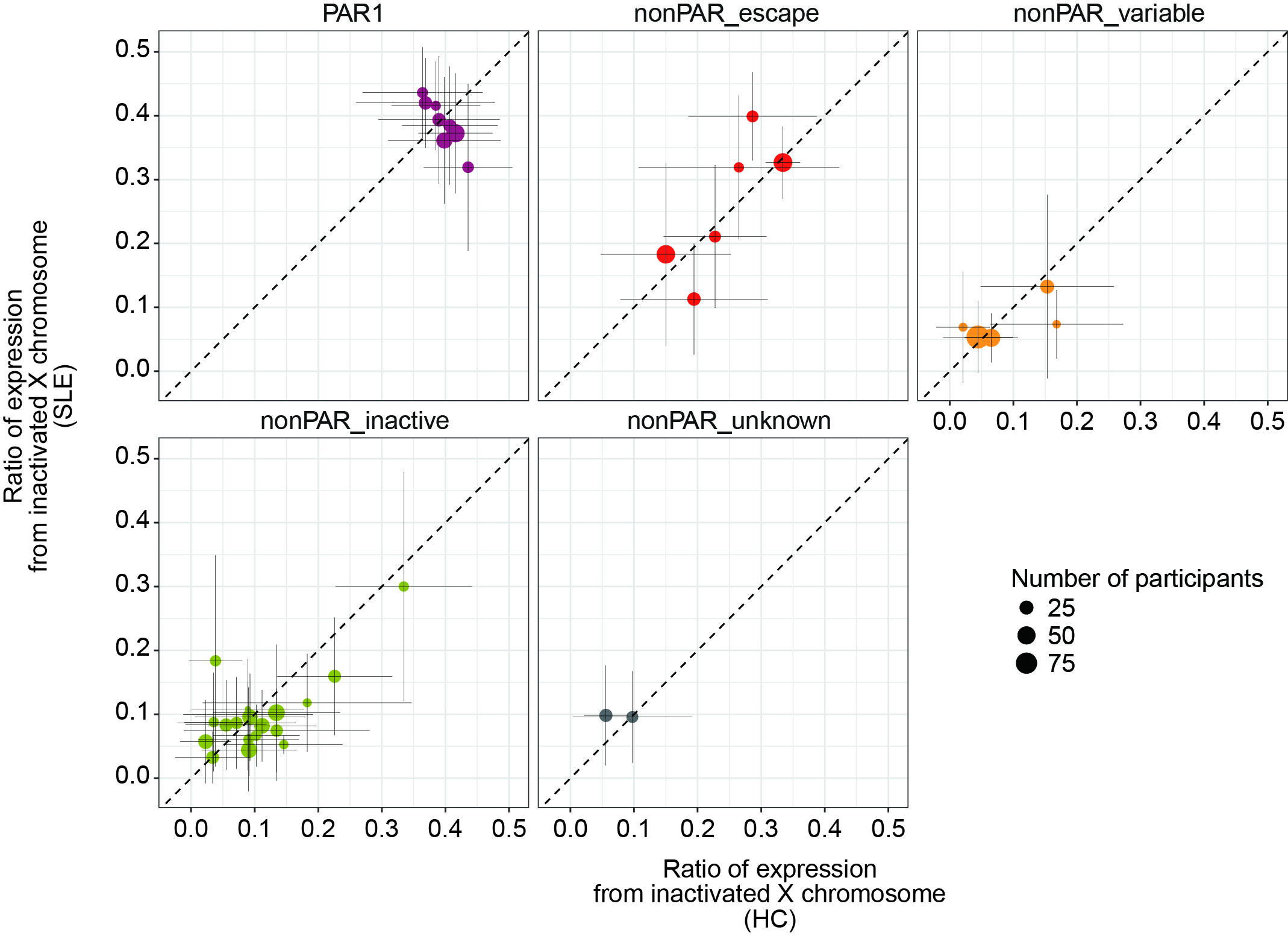
Results: DEG analysis for each immune cell type revealed that degree of female-biased expression of escapee genes was stronger in lymphocytes than in myeloid cells (Figure 1). The scLinaX successfully detected the gene expression from the Xi for the previously reported escapee genes (Figure 2). The scLinaX revealed the stronger degree of escape in lymphocytes than in myeloid cells suggesting that difference in escape makes the heterogeneity of sex-associated differential gene expression across cell types (Figure 1). We also found several genes which had female-biased expression while showing little evidence of the escape in scLinaX analysis, emphasizing the limitation of DEG analysis and the importance of complementary analysis, such as the scLinaX. We evaluated the association between diseases (e.g. SLE) and escape and found little evidence of association, suggesting that abnormal escape in disease conditions might be not common but rather specific for a limited set of reported genes and cell types (Figure 3).
Conclusion: As demonstrated with several public datasets, scLinaX could be applied to a wide variety of scRNA-seq datasets. This study would contribute to expanding the current understanding of the XCI and escape and their contribution to autoimmune diseases.
Y. Tomofuji: None; R. Edahiro: None; Y. Shirai: None; K. Sonehara: None; Q. Wang: None; A. Kumanogoh: None; Y. Okada: None.
Background/Purpose: Understanding the differences in the immune system between sexes is important to elucidate the pathogenesis of autoimmune diseases which are more prevalent in females than males. One of the two X chromosomes of females is silenced through X chromosome inactivation (XCI) to compensate for the difference in the X chromosome dosage between sexes. There exist several genes which 'escape' from XCI, which could contribute to the differential gene expression between sexes. The differences in the escape across cell types are still poorly characterized, especially across immune cell types, even though a large number of immune-related genes are located on the X chromosome. Although previous studies suggested that abnormal escape of a few immune-related genes could contribute to autoimmune diseases, systematic evaluation of the association between escape and autoimmune diseases has not been conducted.
Methods: We investigated the escape across immune cell types utilizing the largest scale scRNA-seq datasets
(1) ~1,000,000 peripheral blood mononuclear cells (PBMCs) from 489 East Asian participants (Asian Immune Diversity Network), (2) ~1,000,000 PBMCs from 147 Japanese (72 COVID19 patients and 75 healthy participants) (3) ~1,000,000 PBMCs from 162 systematic lupus erythematosus patients and 99 healthy participants including East Asian and European.
We performed differential expression gene (DEG) analysis between sexes across immune cell types. In addition, we newly developed a method, single-cell Level inactivated X chromosome mapping (scLinaX), which directly quantifies gene expression from the inactivated X chromosome (Xi). Although the previous studies required plate-based scRNA-seq data which had rich per-cell information at the cost of throughput and cost efficiency for analyzing escape, scLinaX could be applied to the droplet-based scRNA-seq data which are high throughput and widely used.
Results: DEG analysis for each immune cell type revealed that degree of female-biased expression of escapee genes was stronger in lymphocytes than in myeloid cells (Figure 1). The scLinaX successfully detected the gene expression from the Xi for the previously reported escapee genes (Figure 2). The scLinaX revealed the stronger degree of escape in lymphocytes than in myeloid cells suggesting that difference in escape makes the heterogeneity of sex-associated differential gene expression across cell types (Figure 1). We also found several genes which had female-biased expression while showing little evidence of the escape in scLinaX analysis, emphasizing the limitation of DEG analysis and the importance of complementary analysis, such as the scLinaX. We evaluated the association between diseases (e.g. SLE) and escape and found little evidence of association, suggesting that abnormal escape in disease conditions might be not common but rather specific for a limited set of reported genes and cell types (Figure 3).
Conclusion: As demonstrated with several public datasets, scLinaX could be applied to a wide variety of scRNA-seq datasets. This study would contribute to expanding the current understanding of the XCI and escape and their contribution to autoimmune diseases.

Degree of escape tended to be higher in lymphoid cells than in myeloid cells. The plots indicate the result of the comparison of log2 fold changes in the DEG analysis (left) and the ratio of the expression from the inactivated X chromosome (right) between CD4 positive T cells (x-axis) and monocytes (y-axis). The dashed lines indicate y = x.

The scLinaX successfully detected the escape from the inactivated X chromosome for the previously reported escapee genes. A plot indicating the ratio of the gene expression from inactivated X chromosome quantified by scLinaX (y-axis) and the positions on the X chromosome (x-axis). The color of the points indicates whether the genes escape from the X chromosome inactivation or not based on the previous literature. The error bar indicates the standard deviations.

Comparison of the ratio of the gene expression from inactivated X chromosome between healthy control (HC) subjects (x-axis) and systemic lupus erythematosus (SLE) subjects (y-axis). The error bar indicates the standard deviations.
Y. Tomofuji: None; R. Edahiro: None; Y. Shirai: None; K. Sonehara: None; Q. Wang: None; A. Kumanogoh: None; Y. Okada: None.



