Plenary Session
Immunobiology
Session: Plenary II (1579–1584)
1580: CXCL13+ T Cell Differentiation in Systemic Lupus Erythematosus Is Controlled by Opposing Effects of Aryl Hydrocarbon Receptor and Type I Interferon
Monday, November 13, 2023
11:15 AM - 11:25 AM PT
Location: Exhibit Hall A-B
- CL
Calvin Law, PhD (he/him/his)
Northwestern University Feinberg School of Medicine
Chicago, IL, United StatesDisclosure(s): No financial relationships with ineligible companies to disclose
Plenary Presenter(s)
Calvin Law1, Vanessa Wacleche2, Ye Cao3, Arundhati Pillai1, Alice Horisberger4, Sabrina Bracero3, Viktoriya Skidanova3, Zhihan Li3, Ifeoluwakiisi Adejoorin2, Isaac Benque3, Diana Pena Nunez3, Daimon Simmons2, Joshua Keegan4, Lin Chen3, John Sowerby3, Accelerating Medicines Partnership (AMP): RA/SLE2, Elena Massarotti2, Karen Costenbader4, Michael Brenner4, James Lederer4, Judd Hultquist1, Jaehyuk Choi1 and Deepak Rao2, 1Northwestern University Feinberg School of Medicine, Chicago, IL, 2Brigham and Women's Hospital, Boston, MA, 3Brigham and Women's Hospital, Boston, MA, 4Brigham and Women's Hospital and Harvard Medical School, Boston, MA
Background/Purpose: Expansion of B cell-helper T cells including T follicular helper (Tfh) and T peripheral helper (Tph) cells is a prominent feature of systemic lupus erythematosus (SLE). Tfh and Tph cells are marked by high production of the B cell chemoattractant CXCL13; however, regulation of T cell CXCL13 production and the relationship between a CXCL13+ state and other differentiated T cell states remain largely undefined.
Methods: We used mass cytometry to characterize CD4 T cell phenotypes of SLE patients (n=19) and controls (n=19). We used CRISPR screens of primary human CD4 T cells to identify transcription factors that drive core Tph phenotypes. We used transcriptomics (bulk RNA-Seq), epigenetics (ATAC-Seq and CUT&RUN), and functional studies (pharmacological modulators, lentiviral overexpression, reporter cell lines) to characterize mechanisms and pathways that influence CXCL13 production in disease relevant T cells.
Results: Mass cytometry immunophenotyping demonstrated a dramatic imbalance in CD4 T cell phenotypes in SLE patients, with expansion of PD-1+/ICOS+ CXCL13+ T cells and reduction of CD96hi IL-22+ T cells. An arrayed CRISPR screen targeting 80 transcription factors or signaling molecules revealed that deletion of the aryl hydrocarbon receptor (AHR) significantly increased T cell CXCL13 production. Validation experiments confirmed that AHR deletion or pharmacologic inhibition strongly induced T cell CXCL13 production and T cell acquisition of a transcriptomic and epigenetic signature of ex vivo Tph cells. Conversely, AHR activation inhibited a CXCL13 production and a Tph signature while promoting an IL-22+ Th22 cell signature. Luciferase reporter assay using AHR reporter cells show decreased luciferase activity from cells cultured with SLE serum in the presence of AHR agonist. Time coursed transcriptomic analyses of AHR-activated cells combined with unbiased analysis for transcription factor binding sites in AHR CUT&RUN peaks indicated AP-1 transcription factors and AHR co-regulate gene expression. A second CRISPR screen targeting AP-1 family members identified JUN as a potent negative regulator of CXCL13 production and a Tph signature. Further CUT&RUN analyses placed both JUN and AHR at the CXCL13 locus, suggesting direct inhibition of CXCL13 expression. Finally, type I interferon (IFN), a pathogenic driver of SLE, promoted T cell CXCL13 production and a Tph-associated epigenetic signature in part by inhibiting AHR activation and reducing JUN protein expression, while overexpression of JUN inhibited the ability of IFN to induce T cell CXCL13 production.
Conclusion: Our results identify AHR and JUN as potent inhibitors of a Tph cell fate and implicate type I IFN as a key signal in SLE that inhibits AHR and JUN to induce Tph cells.
C. Law: None; V. Wacleche: None; Y. Cao: None; A. Pillai: None; A. Horisberger: None; S. Bracero: None; V. Skidanova: None; Z. Li: None; I. Adejoorin: None; I. Benque: None; D. Pena Nunez: None; D. Simmons: None; J. Keegan: None; L. Chen: None; J. Sowerby: None; A. Medicines Partnership (AMP): RA/SLE: None; E. Massarotti: None; K. Costenbader: Amgen, 2, 5, AstraZeneca, 5, Bristol-Myers Squibb(BMS), 2, Cabaletta, 2, Eli Lilly, 2, Exagen Diagnostics, 5, Gilead, 5, GlaxoSmithKlein(GSK), 2, 5, Janssen, 2, 5; M. Brenner: 4FO Ventures, 2, GlaxoSmithKlein(GSK), 2, Mestag Therapeutics, 2, 11, Third Rock Ventures, 2; J. Lederer: None; J. Hultquist: None; J. Choi: None; D. Rao: AstraZeneca, 2, Bristol-Myers Squibb, 2, 5, GlaxoSmithKlein(GSK), 2, Hifibio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2.
Background/Purpose: Expansion of B cell-helper T cells including T follicular helper (Tfh) and T peripheral helper (Tph) cells is a prominent feature of systemic lupus erythematosus (SLE). Tfh and Tph cells are marked by high production of the B cell chemoattractant CXCL13; however, regulation of T cell CXCL13 production and the relationship between a CXCL13+ state and other differentiated T cell states remain largely undefined.
Methods: We used mass cytometry to characterize CD4 T cell phenotypes of SLE patients (n=19) and controls (n=19). We used CRISPR screens of primary human CD4 T cells to identify transcription factors that drive core Tph phenotypes. We used transcriptomics (bulk RNA-Seq), epigenetics (ATAC-Seq and CUT&RUN), and functional studies (pharmacological modulators, lentiviral overexpression, reporter cell lines) to characterize mechanisms and pathways that influence CXCL13 production in disease relevant T cells.
Results: Mass cytometry immunophenotyping demonstrated a dramatic imbalance in CD4 T cell phenotypes in SLE patients, with expansion of PD-1+/ICOS+ CXCL13+ T cells and reduction of CD96hi IL-22+ T cells. An arrayed CRISPR screen targeting 80 transcription factors or signaling molecules revealed that deletion of the aryl hydrocarbon receptor (AHR) significantly increased T cell CXCL13 production. Validation experiments confirmed that AHR deletion or pharmacologic inhibition strongly induced T cell CXCL13 production and T cell acquisition of a transcriptomic and epigenetic signature of ex vivo Tph cells. Conversely, AHR activation inhibited a CXCL13 production and a Tph signature while promoting an IL-22+ Th22 cell signature. Luciferase reporter assay using AHR reporter cells show decreased luciferase activity from cells cultured with SLE serum in the presence of AHR agonist. Time coursed transcriptomic analyses of AHR-activated cells combined with unbiased analysis for transcription factor binding sites in AHR CUT&RUN peaks indicated AP-1 transcription factors and AHR co-regulate gene expression. A second CRISPR screen targeting AP-1 family members identified JUN as a potent negative regulator of CXCL13 production and a Tph signature. Further CUT&RUN analyses placed both JUN and AHR at the CXCL13 locus, suggesting direct inhibition of CXCL13 expression. Finally, type I interferon (IFN), a pathogenic driver of SLE, promoted T cell CXCL13 production and a Tph-associated epigenetic signature in part by inhibiting AHR activation and reducing JUN protein expression, while overexpression of JUN inhibited the ability of IFN to induce T cell CXCL13 production.
Conclusion: Our results identify AHR and JUN as potent inhibitors of a Tph cell fate and implicate type I IFN as a key signal in SLE that inhibits AHR and JUN to induce Tph cells.

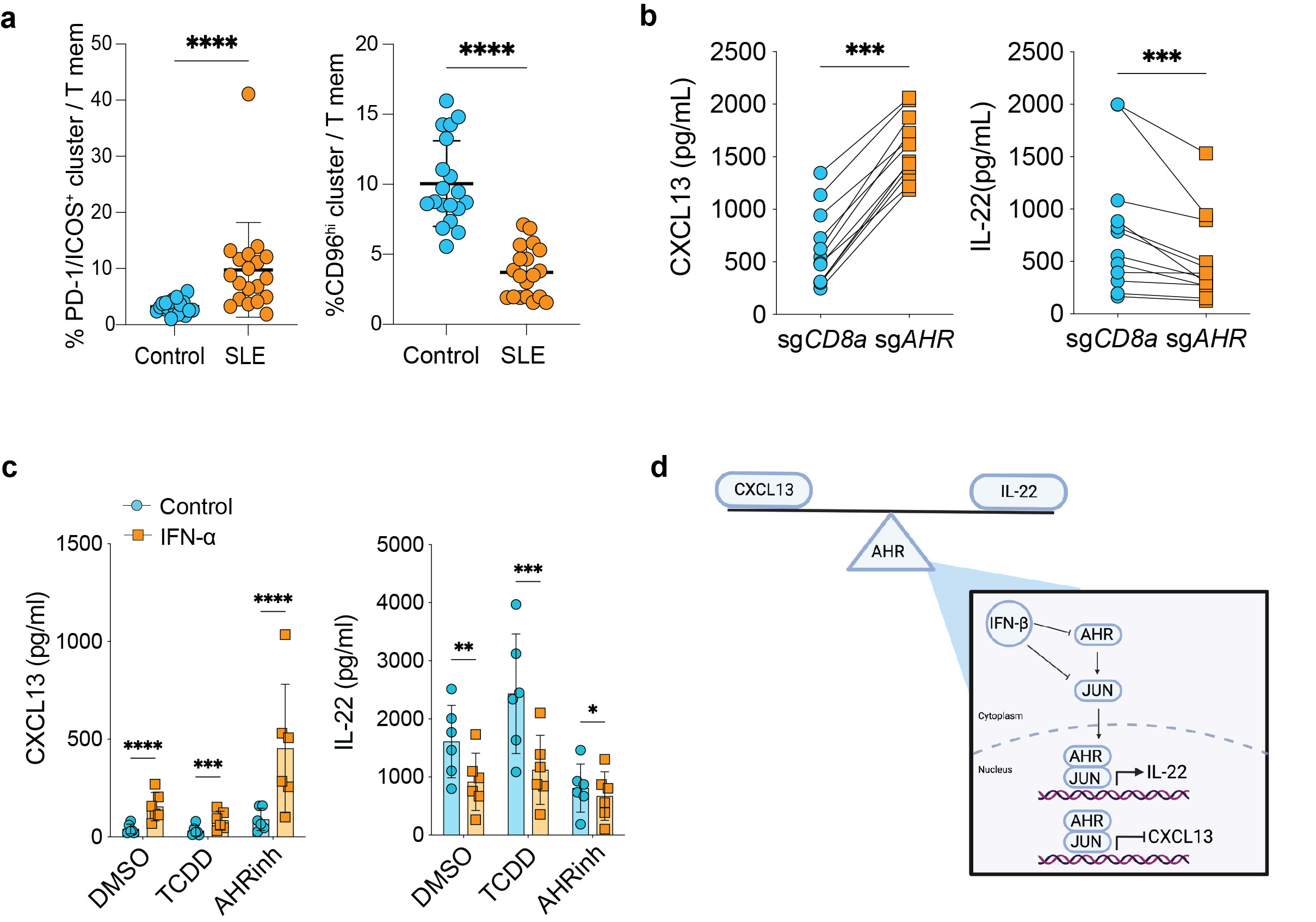
a, Quantification of indicated clusters in SLE patients (n=19) and controls (n=19), P=6.32e-6 for PD-1/ICOS+, P=2.9e-9 for CD96hi.
b, ELISA data for indicated cytokines in supernatants of memory CD4+ T cells nucleofected with sgAHR or sgCD8 control. All cells were cultured with TGF-beta, and each line represents a separate donor (n=12). For CXCL13 P=4.88e-4and IL-22 P=4.88e-4.
c, ELISA for CXCL13 (left) and IL-22 (right) from memory CD4+ T cells stimulated with or without IFN-alpha for 24 hours prior to addition of AHR agonist/inhibitor or DMSO control as indicated (n=6). From left to right, P-value for CXCL13: 8.5e-5, 4.05e-4, 2.9e-5, and for IL-22: 2.35e-3, 8.76e-4, 0.0338.
d, Graphical representation of proposed model in type I IFN regulation of AHR and JUN to promote Tph cells in SLE.
b, ELISA data for indicated cytokines in supernatants of memory CD4+ T cells nucleofected with sgAHR or sgCD8 control. All cells were cultured with TGF-beta, and each line represents a separate donor (n=12). For CXCL13 P=4.88e-4and IL-22 P=4.88e-4.
c, ELISA for CXCL13 (left) and IL-22 (right) from memory CD4+ T cells stimulated with or without IFN-alpha for 24 hours prior to addition of AHR agonist/inhibitor or DMSO control as indicated (n=6). From left to right, P-value for CXCL13: 8.5e-5, 4.05e-4, 2.9e-5, and for IL-22: 2.35e-3, 8.76e-4, 0.0338.
d, Graphical representation of proposed model in type I IFN regulation of AHR and JUN to promote Tph cells in SLE.
C. Law: None; V. Wacleche: None; Y. Cao: None; A. Pillai: None; A. Horisberger: None; S. Bracero: None; V. Skidanova: None; Z. Li: None; I. Adejoorin: None; I. Benque: None; D. Pena Nunez: None; D. Simmons: None; J. Keegan: None; L. Chen: None; J. Sowerby: None; A. Medicines Partnership (AMP): RA/SLE: None; E. Massarotti: None; K. Costenbader: Amgen, 2, 5, AstraZeneca, 5, Bristol-Myers Squibb(BMS), 2, Cabaletta, 2, Eli Lilly, 2, Exagen Diagnostics, 5, Gilead, 5, GlaxoSmithKlein(GSK), 2, 5, Janssen, 2, 5; M. Brenner: 4FO Ventures, 2, GlaxoSmithKlein(GSK), 2, Mestag Therapeutics, 2, 11, Third Rock Ventures, 2; J. Lederer: None; J. Hultquist: None; J. Choi: None; D. Rao: AstraZeneca, 2, Bristol-Myers Squibb, 2, 5, GlaxoSmithKlein(GSK), 2, Hifibio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2.



