Poster Session A
Genetics, genomics and proteomics
Session: (0013–0039.5) Genetics, Genomics & Proteomics Poster
0021: Shared Genetic Susceptibility Between Rheumatoid Arthritis and Bipolar Disorder: Analyses from Genome-Wide Association Studies
Sunday, November 12, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall

Jiayi Zheng, MD
The Wright Center for Graduate Medical Education
Scranton, PA, United StatesDisclosure information not submitted.
Abstract Poster Presenter(s)
Jiayi Zheng1, Ruoning Ni2, Nevena Barjaktarovic3 and Yiming Luo4, 1The Wright Center for Graduate Medical Education, Scranton, PA, 2University of Iowa Hospitals and Clinics, Coralville, IA, 3The Wright Center for Community Health, South Abington Township, PA, 4Division of Rheumatology, Department of Medicine, Columbia University Irving Medical Center, New York, NY
Background/Purpose: Rheumatoid arthritis (RA) has been associated with bipolar disorder (BD) in epidemiological studies, including a recent Mendelian randomization analysis. This study aims to prioritize candidate genes that contribute to the shared genetic susceptibility between these two conditions, using data from genome-wide association studies (GWAS).
Methods: We obtained the summary statistics of the most recent GWAS in RA (PMID: 36333501, 22,350 cases and 74,823 controls) and BD (PMID: 34002096, 41,917 cases and 371,549 controls) of European ancestry. First, we performed linkage disequilibrium score regression (LDSC) to estimate the genetic correlation between the two conditions. We then conducted a cross-trait GWAS meta-analysis in RA and BD to identify shared genetic loci, using a fixed-effect model with METAL software. An additional analysis was carried out reversing the effect direction of BD to detect genetically associated loci with opposite effects. Next, we performed Bayesian genetic colocalization analysis between RA and BD in loci that were significant in the cross-trait GWAS meta-analysis (p < 5x10-8) to predict whether the shared genetic susceptibility was due to the same causal single-nucleotide polymorphisms (SNPs). To evaluate the functional consequences of the discovered signals, loci that were significant in cross-trait meta-analysis and predicted to share the causal SNPs (pp.H4 > 70%) were selected for colocalization with tissue-specific expression quantitative trait loci (eQTL) from the Genotype-Tissue Expression (GTEx), as well as plasma-based protein quantitative trait loci (pQTL) from the Atherosclerosis Risk in Communities (ARIC) dataset. We prioritized genes that colocalized with QTL signals of tissues relevant to RA and BD.
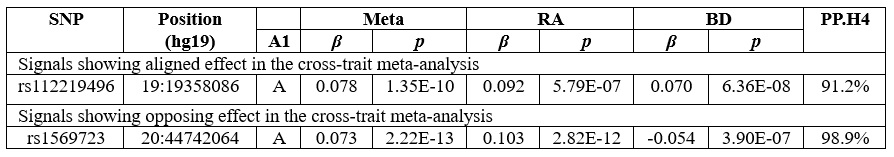
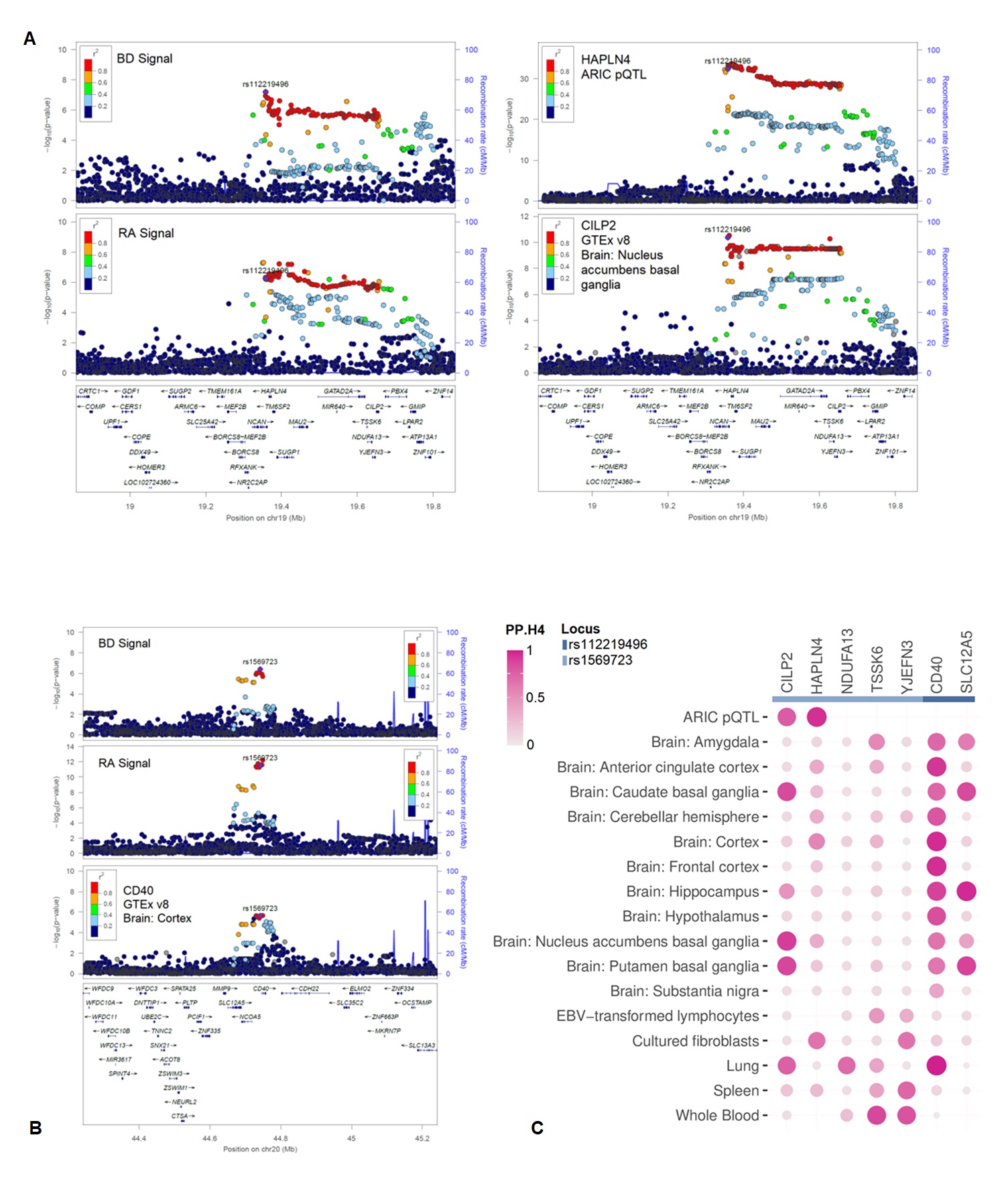
Results: Our analysis showed a modest negative genetic correlation between the two conditions (rg = - 0.1183, se = 0.0315, p = 0.0002). Two genomic loci, near rs112219496 (same effect direction, meta-analysis p = 1.35 x 10-10, pp.H4 = 91.2%) and rs1569723 (opposite effect direction, meta-analysis p-value = 2.22 x 10-13, pp.H4 = 98.9%), were significant in the cross-trait GWAS meta-analysis and colocalized between RA and BD (Table 1). The genomic locus near rs112219496 colocalized with pQTL and eQTLs for CILP2 in plasma, lung, and brain tissues. The genomic locus near rs1569723 colocalized with eQTLs for CD40 in the lung and brain tissues (Figure 1).
Conclusion: This study uncovers a complex relationship between the genetic susceptibilities of RA and BD. By implementing post-GWAS analyses, we identified CILP2 and CD40 as being associated with both RA and BD. Interestingly, CILP2 exhibited the same effect direction in both conditions, while CD40 demonstrated the opposite effect direction.
J. Zheng: None; R. Ni: None; N. Barjaktarovic: None; Y. Luo: None.
Background/Purpose: Rheumatoid arthritis (RA) has been associated with bipolar disorder (BD) in epidemiological studies, including a recent Mendelian randomization analysis. This study aims to prioritize candidate genes that contribute to the shared genetic susceptibility between these two conditions, using data from genome-wide association studies (GWAS).
Methods: We obtained the summary statistics of the most recent GWAS in RA (PMID: 36333501, 22,350 cases and 74,823 controls) and BD (PMID: 34002096, 41,917 cases and 371,549 controls) of European ancestry. First, we performed linkage disequilibrium score regression (LDSC) to estimate the genetic correlation between the two conditions. We then conducted a cross-trait GWAS meta-analysis in RA and BD to identify shared genetic loci, using a fixed-effect model with METAL software. An additional analysis was carried out reversing the effect direction of BD to detect genetically associated loci with opposite effects. Next, we performed Bayesian genetic colocalization analysis between RA and BD in loci that were significant in the cross-trait GWAS meta-analysis (p < 5x10-8) to predict whether the shared genetic susceptibility was due to the same causal single-nucleotide polymorphisms (SNPs). To evaluate the functional consequences of the discovered signals, loci that were significant in cross-trait meta-analysis and predicted to share the causal SNPs (pp.H4 > 70%) were selected for colocalization with tissue-specific expression quantitative trait loci (eQTL) from the Genotype-Tissue Expression (GTEx), as well as plasma-based protein quantitative trait loci (pQTL) from the Atherosclerosis Risk in Communities (ARIC) dataset. We prioritized genes that colocalized with QTL signals of tissues relevant to RA and BD.
Results: Our analysis showed a modest negative genetic correlation between the two conditions (rg = - 0.1183, se = 0.0315, p = 0.0002). Two genomic loci, near rs112219496 (same effect direction, meta-analysis p = 1.35 x 10-10, pp.H4 = 91.2%) and rs1569723 (opposite effect direction, meta-analysis p-value = 2.22 x 10-13, pp.H4 = 98.9%), were significant in the cross-trait GWAS meta-analysis and colocalized between RA and BD (Table 1). The genomic locus near rs112219496 colocalized with pQTL and eQTLs for CILP2 in plasma, lung, and brain tissues. The genomic locus near rs1569723 colocalized with eQTLs for CD40 in the lung and brain tissues (Figure 1).
Conclusion: This study uncovers a complex relationship between the genetic susceptibilities of RA and BD. By implementing post-GWAS analyses, we identified CILP2 and CD40 as being associated with both RA and BD. Interestingly, CILP2 exhibited the same effect direction in both conditions, while CD40 demonstrated the opposite effect direction.

Table 1: Cross-trait meta-analysis of RA and BD and colocalization results of the regions +/- 500 kb of lead SNPs identified in cross-trait meta-analysis.

Figure 1: (A) LocusZoom plots of the region near rs112219496 with RA, BD, pQTL for HAPLN4 and eQTL for CILP2. (B) LocusZoom plots of the region near rs1569723 with RA, BD and eQTL for CD40. (C) Colocalization analysis with pQTLs and eQTLs across various tissues; each row represents a tissue, and each column denotes a gene. The color gradient from violet red to grey signifies the probability of GWAS loci and eQTLs sharing the same causal variant (PPH4).
J. Zheng: None; R. Ni: None; N. Barjaktarovic: None; Y. Luo: None.



