Abstract Session
Periodic fever syndromes, autoinflammatory diseases, Still’s disease and MAS/HLH
Session: Abstracts: Miscellaneous Rheumatic & Inflammatory Diseases II (2515–2520)
2515: Machine Learning-Based Stratification of Mixed Connective Tissue Disease Using Immunophenotyping Data from Patients with Related Autoimmune Diseases
Tuesday, November 14, 2023
4:00 PM - 4:10 PM PT
Location: Room 6E/6D
- SI
Presenting Author(s)
Shinji Izuka1, Toshihiko Komai1, Takahiro Itamiya1, Mineto Ota2, Saeko Yamada1, Yasuo Nagafuchi2, Hirofumi Shoda1, Kosuke Matsuki3, Kazuhiko Yamamoto4, Tomohisa Okamura5 and Keishi Fujio1, 1Department of Allergy and Rheumatology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan, 2Department of Allergy and Rheumatology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan, Department of Functional Genomics and Immunological Diseases, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan, 3Research Division, Chugai Pharmaceutical Co., Ltd., Yokohama City, Japan, 4Department of Allergy and Rheumatology, Graduate School of Medicine, The University of Tokyo and Laboratory for Autoimmune Diseases, Center for Integrative Medical Sciences, RIKEN, Yokohama, Japan, 5Department of Allergy and Rheumatology, Graduate School of Medicine, The University of Tokyo and Department of Functional Genomics and Immunological Diseases, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
Background/Purpose: Mixed connective tissue disease (MCTD) is a heterogenous autoimmune disorder with overlapping clinical features of systemic lupus erythematosus (SLE), polymyositis/dermatomyositis, and systemic sclerosis (SSc). Despite its unique clinical characteristics, some patients may develop other rheumatic diseases during a follow-up period [1]. To better understand the heterogeneity of MCTD and to stratify the patients, we developed machine learning models using immunophenotyping as a reference for SLE, idiopathic inflammatory myopathy (IIM), and SSc and identify differences in symptoms and transcriptome among the subgroups of MCTD.
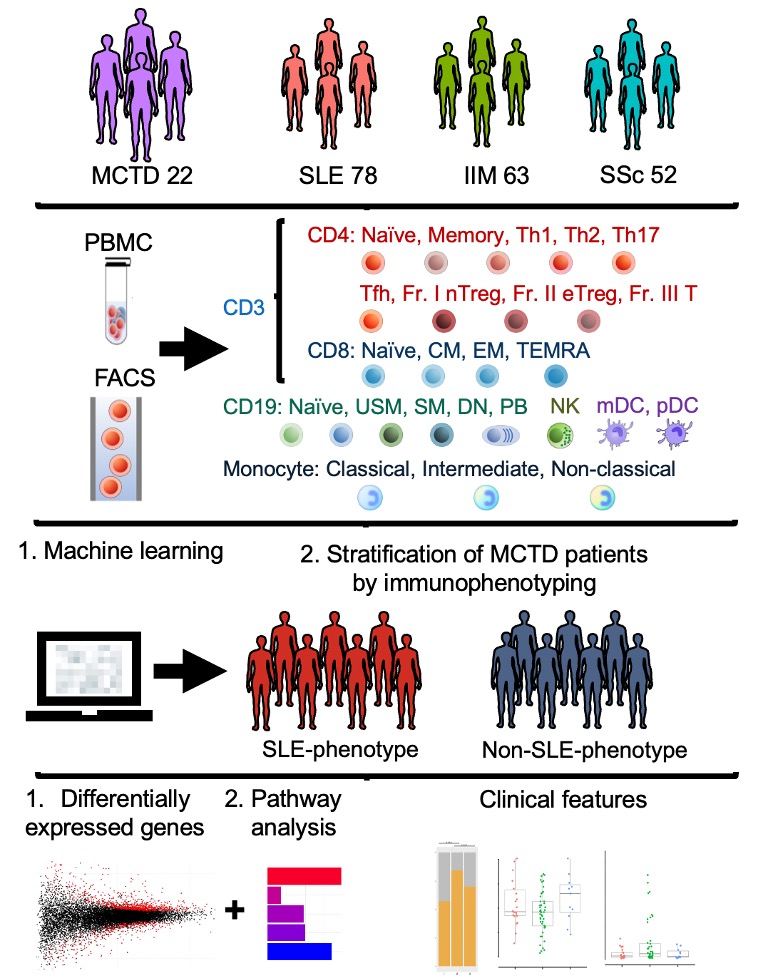
Methods: We employed our large-scale database, Immune Cell Gene Expression Atlas from the University of Tokyo (ImmuNexUT) [2], to analyze the immunophenotyping (24 subsets) of patients with MCTD alongside SLE, IIM, and SSc. By utilizing the immunophenotyping data of SLE, IIM, and SSc, we developed machine learning models such as random forest, neural network, etc., to stratify MCTD patients. Following the stratification of the MCTD patients, we performed a transcriptome analysis, including a differentially expressed gene (DEG) analysis and pathway analysis. Finally, we compared the clinical features of the subgroup of MCTD patients (Figure 1).
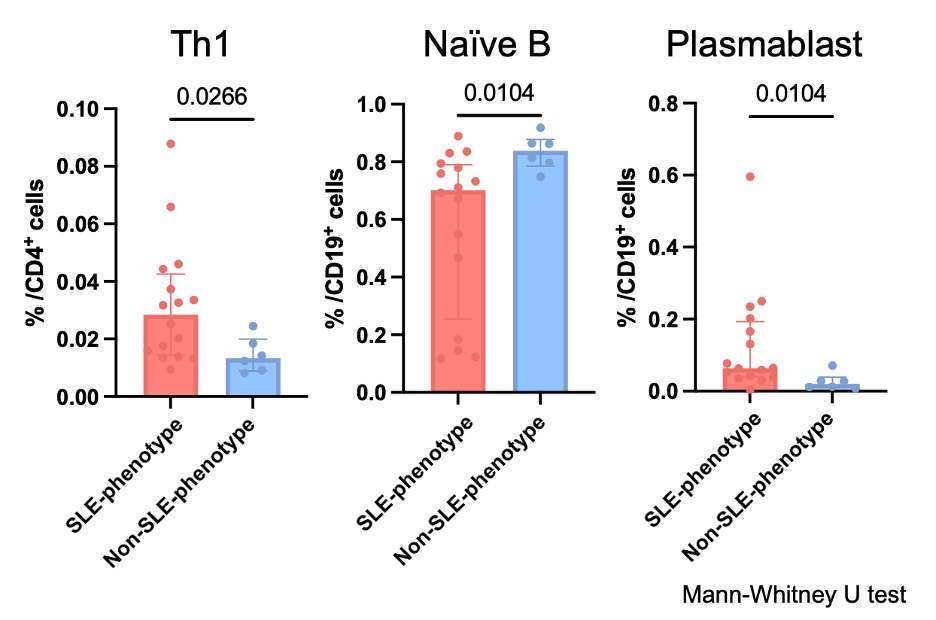
Results: We enrolled a total of 215 patients with autoimmune diseases, consisting of 22 MCTD, 78 SLE, 63 IIM, and 52 SSc patients, and collected the data of immunophenotyping, bulk RNA-sequence on peripheral mononuclear blood cells, and clinical characteristics. We constructed machine learning models to classify patients with SLE, IIM, and SSc based on the immunophenotyping and applied these models to the MCTD patients. Of the 22 patients with MCTD, 16 were classified as SLE-phenotype, and 6 were classified as non-SLE-phenotype (4 IIM-phenotype and 2 SSc-phenotype). Among the MCTD patients, SLE-phenotype patients had significantly higher proportions of Th1, naïve B cells, and plasmablast (p=0.03, 0.01, and 0.01, respectively) (Figure 2). Regarding clinical features, the proportions of having the SLE symptoms such as lymphadenopathy, malar rash, serositis, and cytopenia were significantly higher in SLE-phenotype patients (87.5% vs 33.3%, p=0.025). The number of DEGs across the cell types between SLE patients and MCTD patients (SLE-phenotype and non-SLE-phenotype) was comparable.
Conclusion: Our study suggested the potential stratification of MCTD patients based on their immunophenotyping. This approach may help distinguish clinical phenotypes in MCTD.
[1] Reiseter S et al, Arthritis Res Ther 2017
[2] Ota M et al, Cell 2022
S. Izuka: Eisai, 6; T. Komai: Amgen, 6, Asahi Kasei, 6, Chugai, 5, 6, Daiichi-Sankyo, 6, Eisai, 6, Eli Lilly, 1, 6, GlaxoSmithKlein(GSK), 5, 6, Janssen, 6, Novartis, 6, Tanabe Mitsubishi, 6; T. Itamiya: Chugai Pharmaceutical Co., Ltd., 5; M. Ota: Chugai Pharmaceutical., 12, belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical.; S. Yamada: Asahi Kasei, 6, Chugai Pharmaceutical., 6, Pfizer, 6; Y. Nagafuchi: AbbVie/Abbott, 6, Bristol-Myers Squibb(BMS), 5, 6, Chugai Pharmaceutical., 12, belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical., GlaxoSmithKlein(GSK), 5, Novartis, 6; H. Shoda: AbbVie/Abbott, 6, Asahi Kasei, 6, Astellas, 6, AstraZeneca, 6, Boehringer-Ingelheim, 6, Bristol-Myers Squibb(BMS), 6, Chugai Pharmaceutical., 6, Daiichi-Sankyo, 6, Eisai, 6, Eli Lilly, 6, Gilead, 6, GlaxoSmithKline, 6, Jansen, 6, Novartis, 6, Pfizer, 6, Sanofi, 6, Taisho Pharmaceutical, 6, Takeda, 6; K. Matsuki: Chugai Pharmaceutical., 3; K. Yamamoto: AbbVie, 6, Pfizer Japan Inc, 12, Outsourcing contract, RegCell, 1, Sun Pharmaceutical Industries Ltd, 6; T. Okamura: Chugai Pharmaceutical., 12, belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical.; K. Fujio: AbbVie/Abbott, 6, Asahi Kasei, 5, 6, Astellas, 6, AstraZeneca, 6, Ayumi, 6, Bristol-Myers Squibb(BMS), 5, 6, Chugai Pharmaceutical., 5, 6, Daiichi-Sankyo, 6, Eisai, 5, 6, Eli Lilly, 5, 6, Janssen, 6, Novartis, 6, Ono, 6, Pfizer, 6, Sanofi, 6, Tanabe Mitsubishi, 5, 6, Tsumura, 5.
Background/Purpose: Mixed connective tissue disease (MCTD) is a heterogenous autoimmune disorder with overlapping clinical features of systemic lupus erythematosus (SLE), polymyositis/dermatomyositis, and systemic sclerosis (SSc). Despite its unique clinical characteristics, some patients may develop other rheumatic diseases during a follow-up period [1]. To better understand the heterogeneity of MCTD and to stratify the patients, we developed machine learning models using immunophenotyping as a reference for SLE, idiopathic inflammatory myopathy (IIM), and SSc and identify differences in symptoms and transcriptome among the subgroups of MCTD.
Methods: We employed our large-scale database, Immune Cell Gene Expression Atlas from the University of Tokyo (ImmuNexUT) [2], to analyze the immunophenotyping (24 subsets) of patients with MCTD alongside SLE, IIM, and SSc. By utilizing the immunophenotyping data of SLE, IIM, and SSc, we developed machine learning models such as random forest, neural network, etc., to stratify MCTD patients. Following the stratification of the MCTD patients, we performed a transcriptome analysis, including a differentially expressed gene (DEG) analysis and pathway analysis. Finally, we compared the clinical features of the subgroup of MCTD patients (Figure 1).
Results: We enrolled a total of 215 patients with autoimmune diseases, consisting of 22 MCTD, 78 SLE, 63 IIM, and 52 SSc patients, and collected the data of immunophenotyping, bulk RNA-sequence on peripheral mononuclear blood cells, and clinical characteristics. We constructed machine learning models to classify patients with SLE, IIM, and SSc based on the immunophenotyping and applied these models to the MCTD patients. Of the 22 patients with MCTD, 16 were classified as SLE-phenotype, and 6 were classified as non-SLE-phenotype (4 IIM-phenotype and 2 SSc-phenotype). Among the MCTD patients, SLE-phenotype patients had significantly higher proportions of Th1, naïve B cells, and plasmablast (p=0.03, 0.01, and 0.01, respectively) (Figure 2). Regarding clinical features, the proportions of having the SLE symptoms such as lymphadenopathy, malar rash, serositis, and cytopenia were significantly higher in SLE-phenotype patients (87.5% vs 33.3%, p=0.025). The number of DEGs across the cell types between SLE patients and MCTD patients (SLE-phenotype and non-SLE-phenotype) was comparable.
Conclusion: Our study suggested the potential stratification of MCTD patients based on their immunophenotyping. This approach may help distinguish clinical phenotypes in MCTD.
[1] Reiseter S et al, Arthritis Res Ther 2017
[2] Ota M et al, Cell 2022

Figure 1. Workflow of this study

Figure 2. Comparison of immunophenotyping and clinical features of MCTD patients between SLE-phenotype and non-SLE-phenotype patients.
S. Izuka: Eisai, 6; T. Komai: Amgen, 6, Asahi Kasei, 6, Chugai, 5, 6, Daiichi-Sankyo, 6, Eisai, 6, Eli Lilly, 1, 6, GlaxoSmithKlein(GSK), 5, 6, Janssen, 6, Novartis, 6, Tanabe Mitsubishi, 6; T. Itamiya: Chugai Pharmaceutical Co., Ltd., 5; M. Ota: Chugai Pharmaceutical., 12, belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical.; S. Yamada: Asahi Kasei, 6, Chugai Pharmaceutical., 6, Pfizer, 6; Y. Nagafuchi: AbbVie/Abbott, 6, Bristol-Myers Squibb(BMS), 5, 6, Chugai Pharmaceutical., 12, belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical., GlaxoSmithKlein(GSK), 5, Novartis, 6; H. Shoda: AbbVie/Abbott, 6, Asahi Kasei, 6, Astellas, 6, AstraZeneca, 6, Boehringer-Ingelheim, 6, Bristol-Myers Squibb(BMS), 6, Chugai Pharmaceutical., 6, Daiichi-Sankyo, 6, Eisai, 6, Eli Lilly, 6, Gilead, 6, GlaxoSmithKline, 6, Jansen, 6, Novartis, 6, Pfizer, 6, Sanofi, 6, Taisho Pharmaceutical, 6, Takeda, 6; K. Matsuki: Chugai Pharmaceutical., 3; K. Yamamoto: AbbVie, 6, Pfizer Japan Inc, 12, Outsourcing contract, RegCell, 1, Sun Pharmaceutical Industries Ltd, 6; T. Okamura: Chugai Pharmaceutical., 12, belong to the Social Cooperation Program, Department of Functional Genomics and Immunological Diseases, supported by Chugai Pharmaceutical.; K. Fujio: AbbVie/Abbott, 6, Asahi Kasei, 5, 6, Astellas, 6, AstraZeneca, 6, Ayumi, 6, Bristol-Myers Squibb(BMS), 5, 6, Chugai Pharmaceutical., 5, 6, Daiichi-Sankyo, 6, Eisai, 5, 6, Eli Lilly, 5, 6, Janssen, 6, Novartis, 6, Ono, 6, Pfizer, 6, Sanofi, 6, Tanabe Mitsubishi, 5, 6, Tsumura, 5.



