Abstract Session
Spondyloarthritis (SpA) including psoriatic arthritis (PsA)
Session: Abstracts: Spondyloarthritis Including Psoriatic Arthritis – Basic Science (2437–2442)
2439: Proteomic and Genomic Profiling of Plasma Exosomes from Ankylosing Spondylitis Patients
Tuesday, November 14, 2023
2:30 PM - 2:40 PM PT
Location: Room 30D-E
- FT
Fataneh Tavasolian, PhD
Krembil Research Institute - the University Health Network
Toronto, ON, CanadaDisclosure information not submitted.
Presenting Author(s)
Fataneh Tavasolian1, starlee lively2, Chiara Pastrello3, Michael Tang2, Melissa Lim2, Addison Pacheco2, Zoya Qaiyum2, Enoch Yau2, Zeynep Baskurt4, Igor Jurisica5, Mohit Kapoor6 and Robert Inman2, 1Krembil Research Institute, University Health Network, Toronto, ON, Canada, 2University Health Network, Toronto, ON, Canada, 3Osteoarthritis Research Program, Division of Orthopaedic Surgery, Schroeder Arthritis Institute and Data Science Discovery Centre for Chronic diseases, Krembil Research Institute, Toronto, ON, Canada, 4Department of Biostatistics, University Health Network, Toronto, ON, Canada, 5Schroeder Arthritis Institute, Krembil Research Institute and Departments of Medical Biophysics and Computer Science and Faculty of Dentistry, University of Toronto and Institute of Neuroimmunology, Slovak Academy of Sciences, Bratislava, Slovakia, 6Division of Orthopaedics, Osteoarthritis Research Program, Schroeder Arthritis Institute, and Krembil Research Institute, University Health Network, Toronto, ON, Canada
Background/Purpose: Recent advances in understanding the biology of Ankylosing Spondylitis (AS) using innovative genomic and proteomic approaches offer the opportunity to address current challenges in AS diagnosis and management. Altered expression of genes, microRNAs (miRNAs), or proteins may contribute to immune dysregulation and play a significant role in the onset and persistence of inflammation in AS. The ability of exosomes to transport miRNAs across cells and alter the phenotype of recipient cells has implicated exosomes in perpetuating inflammation in AS. This study reports the first proteomic and miRNA profiling of plasma-derived exosomes in AS using comprehensive computational biology analysis.
Methods: Plasma samples from AS patients and healthy controls (HC) were isolated via ultracentrifugation and subjected to extracellular vesicle (EV) flow cytometry analysis to characterize exosome surface markers by a multiplex immunocapture assay. Cytokine profiling of plasma-derived exosomes and cell culture supernatants was performed. Next-generation sequencing was used to identify miRNA populations in exosomes enriched from plasma fractions. CD4+T cells were sorted, and the frequency and proliferation of CD4+T cell subsets were analyzed after treatment with AS-exosomes using flow cytometry.
Results: The expression of exosome-marker proteins CD63 and CD81 was elevated in the AS patients compared to HC (q < 0.05). Cytokine profiling in plasma-derived AS-exosomes demonstrated downregulation of interleukin (IL)-8 and IL-10 (q < 0.05). AS-exosomes co-cultured with HC CD4+ T cells induced significant upregulation of IFNα2 and IL-33 (q < 0.05). Exosomes from AS patients inhibited the proliferation of regulatory T cells (Treg), suggesting a mechanism for chronically activated T cells in this disease. Culture of CD4+T cells from healthy individuals in the presence of AS-exosomes reduced the proliferation of FOXP3+Treg cells and decreased the frequency of FOXP3+IRF4+Treg cells. miRNA sequencing identified 24 differentially expressed miRNAs found in circulating exosomes of AS patients compared with HC; 22 of which were upregulated, and 2 were downregulated.
Conclusion: Individuals with AS have different immunological and genetic profiles, as determined by evaluating the exosomes of these patients. The inhibitory effect of exosomes on Treg in AS suggests a mechanism contributing to chronically activated T cells in this disease.
F. Tavasolian: None; s. lively: None; C. Pastrello: None; M. Tang: None; M. Lim: None; A. Pacheco: None; Z. Qaiyum: None; E. Yau: None; Z. Baskurt: None; I. Jurisica: None; M. Kapoor: None; R. Inman: AbbVie, 2, Eli Lilly, 2, Janssen, 2, Novartis, 2, Sandoz, 2.
Background/Purpose: Recent advances in understanding the biology of Ankylosing Spondylitis (AS) using innovative genomic and proteomic approaches offer the opportunity to address current challenges in AS diagnosis and management. Altered expression of genes, microRNAs (miRNAs), or proteins may contribute to immune dysregulation and play a significant role in the onset and persistence of inflammation in AS. The ability of exosomes to transport miRNAs across cells and alter the phenotype of recipient cells has implicated exosomes in perpetuating inflammation in AS. This study reports the first proteomic and miRNA profiling of plasma-derived exosomes in AS using comprehensive computational biology analysis.
Methods: Plasma samples from AS patients and healthy controls (HC) were isolated via ultracentrifugation and subjected to extracellular vesicle (EV) flow cytometry analysis to characterize exosome surface markers by a multiplex immunocapture assay. Cytokine profiling of plasma-derived exosomes and cell culture supernatants was performed. Next-generation sequencing was used to identify miRNA populations in exosomes enriched from plasma fractions. CD4+T cells were sorted, and the frequency and proliferation of CD4+T cell subsets were analyzed after treatment with AS-exosomes using flow cytometry.
Results: The expression of exosome-marker proteins CD63 and CD81 was elevated in the AS patients compared to HC (q < 0.05). Cytokine profiling in plasma-derived AS-exosomes demonstrated downregulation of interleukin (IL)-8 and IL-10 (q < 0.05). AS-exosomes co-cultured with HC CD4+ T cells induced significant upregulation of IFNα2 and IL-33 (q < 0.05). Exosomes from AS patients inhibited the proliferation of regulatory T cells (Treg), suggesting a mechanism for chronically activated T cells in this disease. Culture of CD4+T cells from healthy individuals in the presence of AS-exosomes reduced the proliferation of FOXP3+Treg cells and decreased the frequency of FOXP3+IRF4+Treg cells. miRNA sequencing identified 24 differentially expressed miRNAs found in circulating exosomes of AS patients compared with HC; 22 of which were upregulated, and 2 were downregulated.
Conclusion: Individuals with AS have different immunological and genetic profiles, as determined by evaluating the exosomes of these patients. The inhibitory effect of exosomes on Treg in AS suggests a mechanism contributing to chronically activated T cells in this disease.

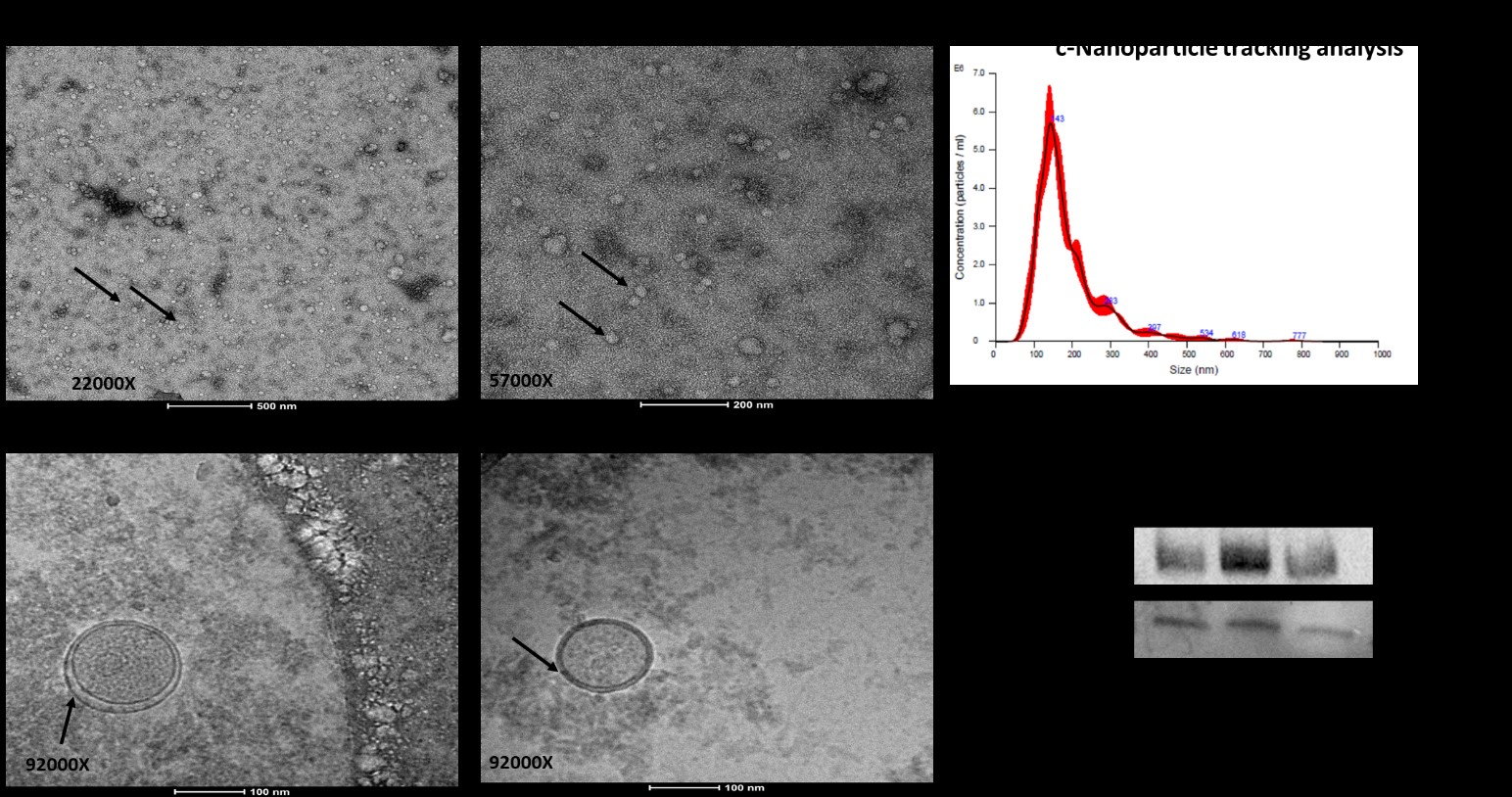
Figure 1. (a) Exosome characteristics with morphology, size, and markers. Exosomes display a round-shaped morphology in negative TEM. It is on 22000x magnification, demonstrating the density of isolated exosomes in plasma (b)and are bilayer and spherical by cryo-TEM imaging. (c) Exosome sizes can be determined using nanoparticle tracking. NTA was used to examine the exosome size and concentration, and the results showed that the exosomes had an average size of 147 nm. (d) Expression of the exosomal markers CD63 and CD9 was confirmed with Western blot.
F. Tavasolian: None; s. lively: None; C. Pastrello: None; M. Tang: None; M. Lim: None; A. Pacheco: None; Z. Qaiyum: None; E. Yau: None; Z. Baskurt: None; I. Jurisica: None; M. Kapoor: None; R. Inman: AbbVie, 2, Eli Lilly, 2, Janssen, 2, Novartis, 2, Sandoz, 2.



