Abstract Session
Infection-related rheumatic syndromes
Session: Abstracts: Infection-related Rheumatic Disease (1669–1674)
1671: Use of Metagenomic Microbial Plasma Cell-Free DNA Next-Generation Sequencing Assay in Outpatient Rheumatology Practice
Monday, November 13, 2023
4:30 PM - 4:40 PM PT
Location: Room 6E/6D
- RJ
Presenting Author(s)
Rachel Jenkins, Matthew Samec, Courtney Arment and Matthew Koster, Mayo Clinic, Rochester, MN
Background/Purpose: Metagenomic next-generation sequencing (mNGS) of microbial cell-free DNA (mcfDNA) allows for non-invasive broad-range pathogen detection from plasma. The Karius test (KT) is a commercially available mNGS mcfDNA technology able to evaluate over 1000 pathogens on a single blood draw. Patients presenting with inflammatory syndromes often have overlapping clinical features and in some circumstances, differentiation between autoimmune disease and indolent infection can be challenging. This is of particular importance in patients where infection needs to be excluded prior to the initiation or escalation of immunosuppression. The purpose of this study was to describe the clinical utility of KT in a tertiary outpatient rheumatology practice.
Methods: All patients for which a KT was ordered by a rheumatology provider during outpatient evaluation at Mayo Clinic, Rochester, MN between 7/1/2021 and 12/31/2022 were identified. Demographics, symptoms, and laboratory parameters at time of KT draw were abstracted. Reason for testing was categorized as fever of unknown origin (FUO), atypical presentation of possible rheumatic disease (RD) or assessment of RD flare versus infection. Results were considered negative (-) if no pathogen was listed as above KT threshold for positive. Among positive tests, determination of an organism's clinical relevance was assessed based on whether the listed pathogen was considered associated with the clinical presentation for which the KT was drawn and/or resulted in initiation (or modification) of anti-microbial therapy to address the detected organism. Records of patients with (-) KT were reviewed for a minimum of 3 months after KT draw to determine if patients with a (-) KT had subsequent identification of pathologic organism by another testing method.
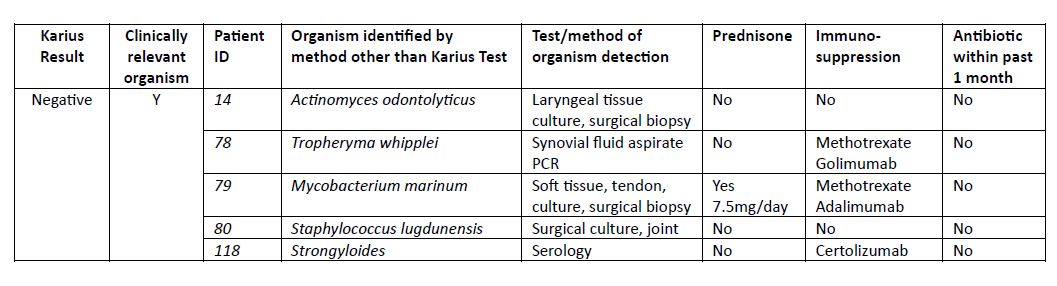
Results: 150 patients with an outpatient KT were identified, 53% female, mean age of 52 years. Reasons for KT were atypical presentation of possible RD (80%), evaluation for flare of RD vs infection (16.7%) and FUO (3.3%). In total 24 (16%) patients had at least one organism listed as detected, among which 25% (6/24) were considered clinically relevant and altered final diagnosis and treatment (Table 2). 126 (84%) patients had a (-) KT of which 5 (4%) were subsequently found to have a clinically relevant infection by other testing (Table 3). Among the 121 patients with (-) KT for which alternative organism was not seen, 55 started and 11 increased immunosuppression after (-) KT; none of which were diagnosed with an infection associated with initial presentation within the subsequent 3 months after the KT test.
Conclusion: This study describes the first large series of mNGS mcfDNA in outpatient rheumatology. Six patients were found to have systemic infections which altered diagnosis and treatment. KT false (-) rate was only 4% and 3 of the 5 cases with false (-) KT required surgical biopsy for organism confirmation. Over 50% of patients with negative KT were started on immunosuppression without subsequent infection identified. Determining which patients best benefit from KT use in rheumatology practice remains to be defined. Further research is needed prior to consideration of widespread use.
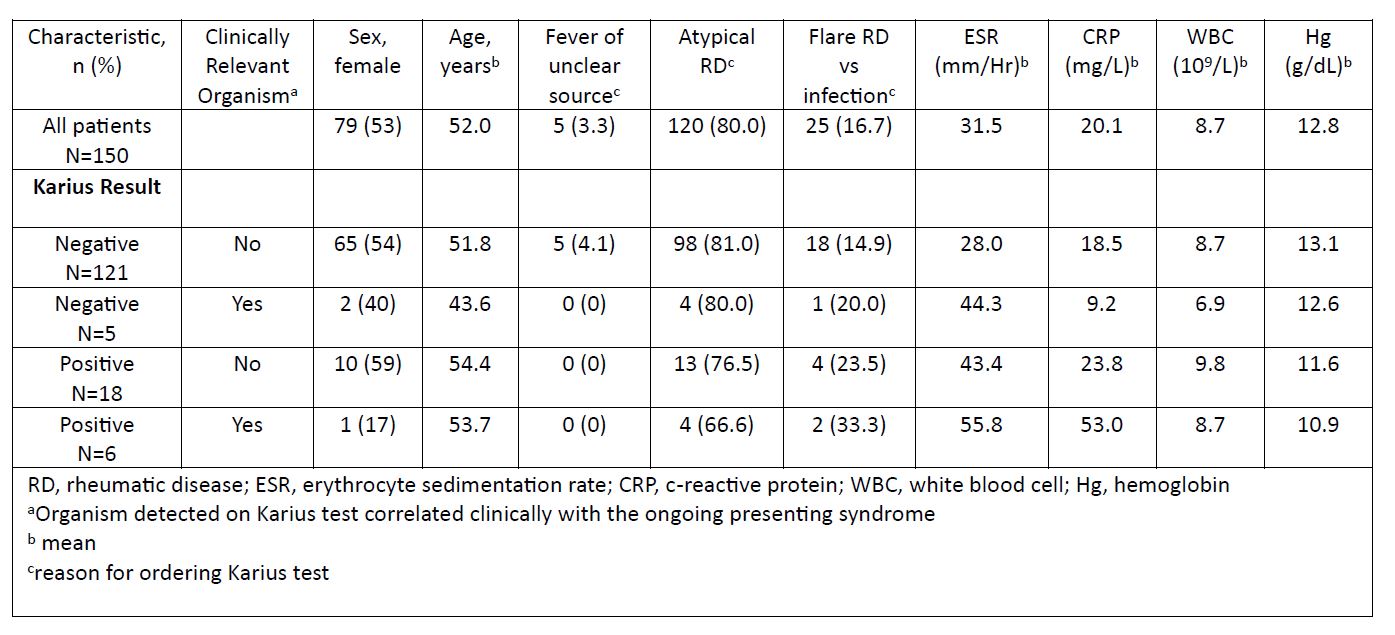
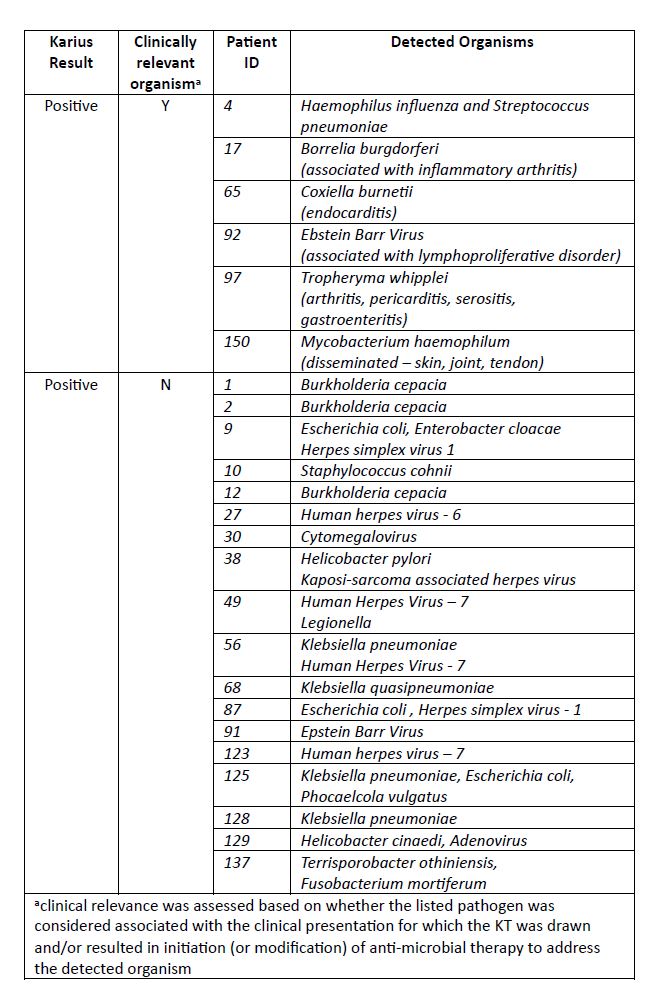
Table 2: Organisms identified among patients with clinically relevant and non-clinically relevant positive Karius test results
R. Jenkins: None; M. Samec: None; C. Arment: None; M. Koster: None.
Background/Purpose: Metagenomic next-generation sequencing (mNGS) of microbial cell-free DNA (mcfDNA) allows for non-invasive broad-range pathogen detection from plasma. The Karius test (KT) is a commercially available mNGS mcfDNA technology able to evaluate over 1000 pathogens on a single blood draw. Patients presenting with inflammatory syndromes often have overlapping clinical features and in some circumstances, differentiation between autoimmune disease and indolent infection can be challenging. This is of particular importance in patients where infection needs to be excluded prior to the initiation or escalation of immunosuppression. The purpose of this study was to describe the clinical utility of KT in a tertiary outpatient rheumatology practice.
Methods: All patients for which a KT was ordered by a rheumatology provider during outpatient evaluation at Mayo Clinic, Rochester, MN between 7/1/2021 and 12/31/2022 were identified. Demographics, symptoms, and laboratory parameters at time of KT draw were abstracted. Reason for testing was categorized as fever of unknown origin (FUO), atypical presentation of possible rheumatic disease (RD) or assessment of RD flare versus infection. Results were considered negative (-) if no pathogen was listed as above KT threshold for positive. Among positive tests, determination of an organism's clinical relevance was assessed based on whether the listed pathogen was considered associated with the clinical presentation for which the KT was drawn and/or resulted in initiation (or modification) of anti-microbial therapy to address the detected organism. Records of patients with (-) KT were reviewed for a minimum of 3 months after KT draw to determine if patients with a (-) KT had subsequent identification of pathologic organism by another testing method.
Results: 150 patients with an outpatient KT were identified, 53% female, mean age of 52 years. Reasons for KT were atypical presentation of possible RD (80%), evaluation for flare of RD vs infection (16.7%) and FUO (3.3%). In total 24 (16%) patients had at least one organism listed as detected, among which 25% (6/24) were considered clinically relevant and altered final diagnosis and treatment (Table 2). 126 (84%) patients had a (-) KT of which 5 (4%) were subsequently found to have a clinically relevant infection by other testing (Table 3). Among the 121 patients with (-) KT for which alternative organism was not seen, 55 started and 11 increased immunosuppression after (-) KT; none of which were diagnosed with an infection associated with initial presentation within the subsequent 3 months after the KT test.
Conclusion: This study describes the first large series of mNGS mcfDNA in outpatient rheumatology. Six patients were found to have systemic infections which altered diagnosis and treatment. KT false (-) rate was only 4% and 3 of the 5 cases with false (-) KT required surgical biopsy for organism confirmation. Over 50% of patients with negative KT were started on immunosuppression without subsequent infection identified. Determining which patients best benefit from KT use in rheumatology practice remains to be defined. Further research is needed prior to consideration of widespread use.

Table 1. Baseline characteristics of Karius testing groups

Table 2: Organisms identified among patients with clinically relevant and non-clinically relevant positive Karius test results

Table 3: Organisms detected by other methodology in patients with negative Karius test
R. Jenkins: None; M. Samec: None; C. Arment: None; M. Koster: None.



