Abstract Session
Spondyloarthritis (SpA) including psoriatic arthritis (PsA)
Session: Abstracts: Spondyloarthritis Including Psoriatic Arthritis – Diagnosis, Manifestations, & Outcomes II: Psoriatic Arthritis (1639–1644)
1643: Exploring the Serum Metabolome for Potential Diagnostic Markers of Psoriatic Arthritis
Monday, November 13, 2023
3:00 PM - 3:10 PM PT
Location: Ballroom 20A
- VC
Vinod Chandran, MD, PhD, MBBS, DM, FRCPC
University of Toronto
Toronto, ON, CanadaDisclosure information not submitted.
Presenting Author(s)
Nikita Looby1, Max Kotlyar2, Chiara Pastrello2, Darshini Ganatra1, Vathany Kulasingam3, Igor Jurisica4 and Vinod Chandran5, 1Schroeder Arthritis Institute, Krembil Research Institute, University Health Network, Toronto, ON, Canada, 2Osteoarthritis Research Program, Division of Orthopaedic Surgery, Schroeder Arthritis Institute and Data Science Discovery Centre for Chronic diseases, Krembil Research Institute, Toronto, ON, Canada, 3Department of Laboratory Medicine and Pathobiology, University of Toronto and Division of Clinical Biochemistry, Laboratory Medicine Program, University Health Network, Toronto, ON, Canada, 4Schroeder Arthritis Institute, Krembil Research Institute and Departments of Medical Biophysics and Computer Science and Faculty of Dentistry, University of Toronto and Institute of Neuroimmunology, Slovak Academy of Sciences, Bratislava, Slovakia, 5Schroeder Arthritis Institute, Krembil Research Institute, University Health Network and Division of Rheumatology, Department of Medicine, University of Toronto, Toronto, ON, Canada
Background/Purpose: Psoriatic arthritis (PsA) is an inflammatory immune-mediated musculoskeletal skin disease that affects approximately 25% of psoriasis patients causing progressive disability. As a heterogeneous disease, with sometimes subtle manifestation, accurate assessment of PsA is difficult. As such, there is a need for early detection diagnostic tests for PsA. A liquid chromatography – high resolution mass spectrometry (LC-HRMS) general untargeted metabolomics analysis following solid phase microextraction (SPME) was applied to serum samples collected from patients with PsA and psoriasis without PsA (PsC), to perform discovery analysis to investigate potential diagnostic markers of PsA.
Methods: Serum samples were obtained from a biobank of carefully phenotyped PsC (n=100) and PsA (n = 101) patients who had no history of cancer, had no active infection nor received previous treatment with biologics. Novel high throughput technique – SPME – was used to prepare all samples simultaneously followed by LC-HRMS analysis.
Data (pre)processing and feature identification was performed using 2 workflows - Compound Discoverer 3.3 and an independent Rscript. Supervised multivariate analysis and various Machine Learning (ML) algorithms including logitboost, adaptive boosting, support vector machine (SVM), logistic regression, and random forest (RF), were used for predictive feature analysis. Area Under the Receiver Operating Characteristic (AUROC) was then used to evaluate the performance of these features. Only features in models with an area under the curve (AUC) of 0.85 or greater were considered as candidate biomarkers.
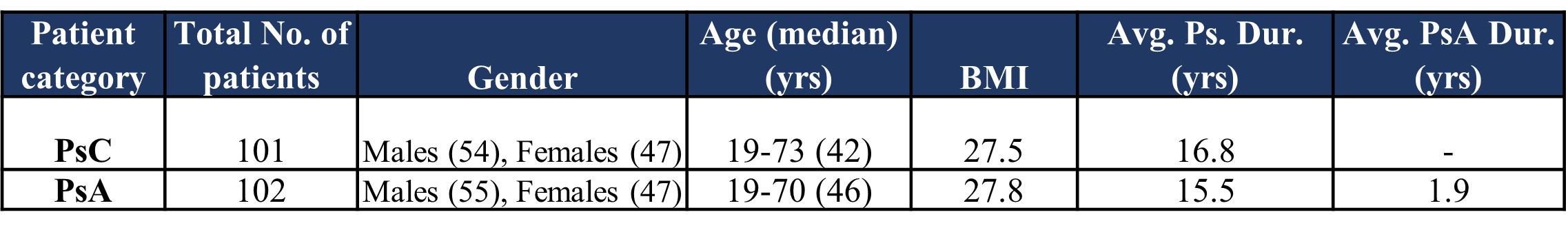
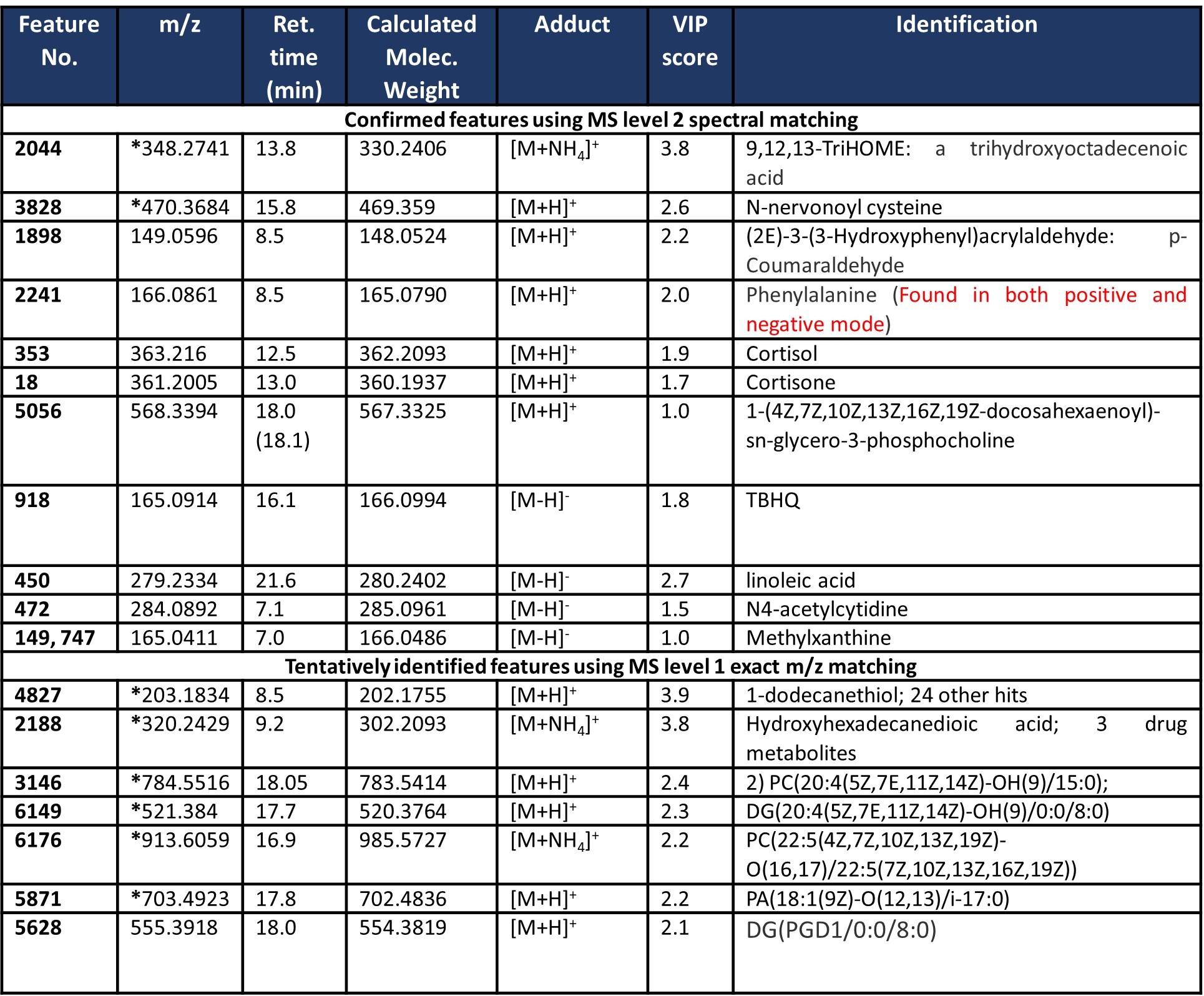
Results: Table 1 provides the demographic and disease characteristics of the included subjects. Table 2 provides a summary of the results from predictive feature analyses. A minimum of ten features and a maximum of eighty features from the adaptive boost models produced an AUC of 0.896 and 0.921 respectively. All other models with feature numbers ranging between twenty to eighty produced AUC between 0.891 and 0.915. Several small molecules could be validated via MS Level 2 spectral database matching. Trihydroxyoctadecenoic acid and N-nervonoyl cysteine contributed significantly to the multivariate supervised analysis and performed well according to high AUROC scores. Interestingly, lipids such as docosahexanoyl-sn-gylcerophosphocholine were identified via MS level 2, but there were several other features that were found to be significant that could only be tentatively identified as glycero-lipids and fatty acids. Results from this metabolomics workflow were integrated with top-down multi-omics data for the same patients. Data integration revealed that confirmed and tentatively identified features like glycero- and phospholipids overlapped differentially expressed pathways from the transcriptome.
Conclusion: SPME-LC-HRMS based untargeted metabolomic analyses have identified small molecules (lipids) with excellent discriminative ability between PsA and PsC. Thus, the development of quantitative targeted assays for these metabolites and subsequent validation may provide diagnostic markers for PsA.
N. Looby: None; M. Kotlyar: None; C. Pastrello: None; D. Ganatra: None; V. Kulasingam: None; I. Jurisica: None; V. Chandran: AbbVie, 1, 5, 6, Amgen, 1, 5, 6, AstraZeneca, 3, Bristol-Myers Squibb (BMS), 1, 6, Eli Lilly, 1, 5, 6, Janssen, 1, 6, Novartis, 1, 1, 6, UCB, 1, 2.
Background/Purpose: Psoriatic arthritis (PsA) is an inflammatory immune-mediated musculoskeletal skin disease that affects approximately 25% of psoriasis patients causing progressive disability. As a heterogeneous disease, with sometimes subtle manifestation, accurate assessment of PsA is difficult. As such, there is a need for early detection diagnostic tests for PsA. A liquid chromatography – high resolution mass spectrometry (LC-HRMS) general untargeted metabolomics analysis following solid phase microextraction (SPME) was applied to serum samples collected from patients with PsA and psoriasis without PsA (PsC), to perform discovery analysis to investigate potential diagnostic markers of PsA.
Methods: Serum samples were obtained from a biobank of carefully phenotyped PsC (n=100) and PsA (n = 101) patients who had no history of cancer, had no active infection nor received previous treatment with biologics. Novel high throughput technique – SPME – was used to prepare all samples simultaneously followed by LC-HRMS analysis.
Data (pre)processing and feature identification was performed using 2 workflows - Compound Discoverer 3.3 and an independent Rscript. Supervised multivariate analysis and various Machine Learning (ML) algorithms including logitboost, adaptive boosting, support vector machine (SVM), logistic regression, and random forest (RF), were used for predictive feature analysis. Area Under the Receiver Operating Characteristic (AUROC) was then used to evaluate the performance of these features. Only features in models with an area under the curve (AUC) of 0.85 or greater were considered as candidate biomarkers.
Results: Table 1 provides the demographic and disease characteristics of the included subjects. Table 2 provides a summary of the results from predictive feature analyses. A minimum of ten features and a maximum of eighty features from the adaptive boost models produced an AUC of 0.896 and 0.921 respectively. All other models with feature numbers ranging between twenty to eighty produced AUC between 0.891 and 0.915. Several small molecules could be validated via MS Level 2 spectral database matching. Trihydroxyoctadecenoic acid and N-nervonoyl cysteine contributed significantly to the multivariate supervised analysis and performed well according to high AUROC scores. Interestingly, lipids such as docosahexanoyl-sn-gylcerophosphocholine were identified via MS level 2, but there were several other features that were found to be significant that could only be tentatively identified as glycero-lipids and fatty acids. Results from this metabolomics workflow were integrated with top-down multi-omics data for the same patients. Data integration revealed that confirmed and tentatively identified features like glycero- and phospholipids overlapped differentially expressed pathways from the transcriptome.
Conclusion: SPME-LC-HRMS based untargeted metabolomic analyses have identified small molecules (lipids) with excellent discriminative ability between PsA and PsC. Thus, the development of quantitative targeted assays for these metabolites and subsequent validation may provide diagnostic markers for PsA.

Table 1: Summary of patient demographics and disease characteristics

Table 2: Confirmed and tentatively identified features of statistical significance. These features were implicated in a cross validated PLS-DA model. Features containing a (*) were also found to perform well (>0.85) using Area Under the Receiver Operating Characteristic (AUROC).
N. Looby: None; M. Kotlyar: None; C. Pastrello: None; D. Ganatra: None; V. Kulasingam: None; I. Jurisica: None; V. Chandran: AbbVie, 1, 5, 6, Amgen, 1, 5, 6, AstraZeneca, 3, Bristol-Myers Squibb (BMS), 1, 6, Eli Lilly, 1, 5, 6, Janssen, 1, 6, Novartis, 1, 1, 6, UCB, 1, 2.



