Abstract Session
Osteoarthritis (OA) and related disorders
Session: Abstracts: Osteoarthritis & Joint Biology – Basic Science (1591–1596)
1593: Combining Single-cell RNA Sequencing and Population-based Studies Reveals Hand Osteoarthritis-associated Chondrocyte Subpopulations and Pathways
Monday, November 13, 2023
2:30 PM - 2:40 PM PT
Location: Room 30D-E
- HL
Hui Li, MD, PhD
Department of Orthopaedics, Xiangya Hospital, Central South University
Changsha, ChinaDisclosure information not submitted.
Presenting Author(s)
Hui Li1, Xiaofeng Jiang1, Yongbing Xiao1, Yuqing Zhang2, Weiya Zhang3, Michael Doherty3, Jacquelyn Nestor4, Changjun Li5, Jing Ye1, Tingting Sha6, Houchen Lyu1, Jie Wei7, Chao Zeng1 and Guanghua Lei1, 1Department of Orthopaedics, Xiangya Hospital, Central South University, Changsha, China, 2Division of Rheumatology, Allergy, and Immunology, Department of Medicine, Massachusetts General Hospital, Boston, MA, 3Academic Rheumatology, Clinical Sciences Building, University of Nottingham, City Hospital, Nottingham, United Kingdom, 4Division of Rheumatology, Allergy, and Immunology, Department of Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA, 5National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China, 6Hunan Key Laboratory of Joint Degeneration and Injury, Changsha, China, 7Health Management Center, Xiangya Hospital Central South University, Changsha, China
Background/Purpose: Hand osteoarthritis (OA) is a common heterogeneous joint disorder with differences in etiology and pathophysiology from the large weight-bearing knee or hip OA. Its pathophysiological mechanism remains largely unexplored, partially because of limited access to clinical sample tissues and lack of animal models. To date, there is no known cure for hand OA, indicating an urgent need for better understanding of the underlying mechanisms to develop appropriate treatment strategies. Here, we aimed to identify hand joint chondrocyte subpopulations and investigate the molecular mechanism of hand OA by performing single-cell RNA sequencing (scRNA-seq) analysis, and verify the findings in two large independent population-based studies.
Methods: We obtained hand interphalangeal joints from five donors who had destructive forearm injury. Using scRNA-seq analysis, we analyzed the cellular composition and subpopulation-specific gene expression of hand articular cartilage, and then compared these features between cartilages from joints with macroscopically confirmed OA (n=5) and those without OA (n=5). We further conducted: (1) a Mendelian randomization study using data from the UK Biobank to investigate the causal association between the key differentially expressed gene and hand OA; and (2) a cross-sectional study using data collected from a community-based observational study, i.e., the Xiangya OA (XO) Study, to examine the association between a serum biomarker (encoded by the key gene) and hand OA.
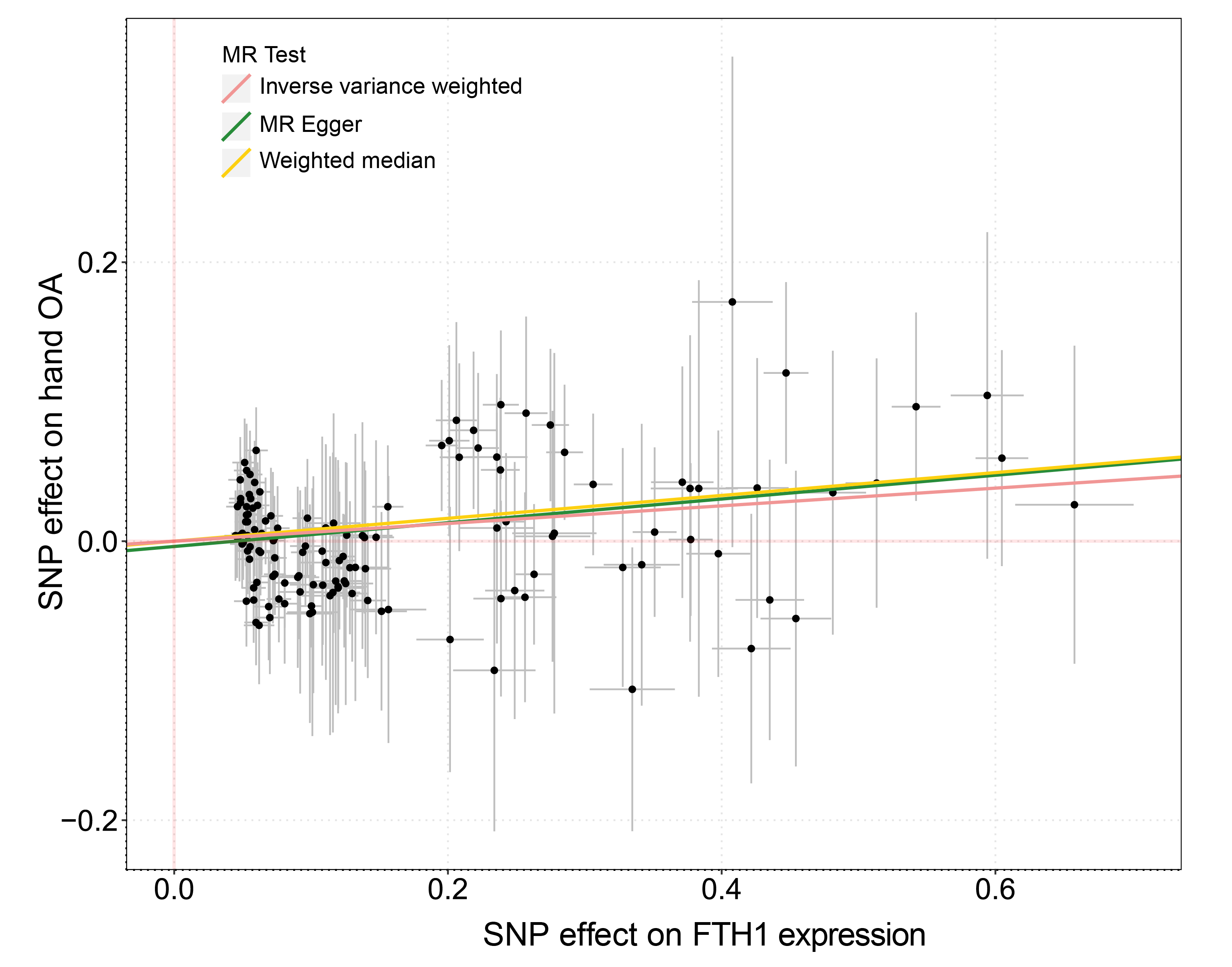
Results: Of 105,142 cells we identified 13 subpopulations, including a novel inflammatory chondrocyte subpopulation that specifically expressed genes related to inflammatory and immune response. Fibrocartilage chondrocytes represented a major source of OA-related proteases and exhibited an extensive alteration of gene expression patterns in OA cartilage compared with non-OA cartilage. Both inflammatory chondrocytes and fibrocartilage chondrocytes showed a trend towards increased numbers in hand OA cartilage. In these two subpopulations from OA cartilage, the ferroptosis pathway was enriched, in which the expression of iron overload-related genes, e.g., FTH1, was elevated. These findings are further validated by two independent population-based studies. Among participants (n=332,668) in the UK Biobank, genetic predisposition to higher expression of FTH1 mRNA significantly increased the risk of hand OA (OR=1.07, 95%CI:1.02-1.11). Among participants (n=1,241) from the XO Study, high levels of serum ferritin (encoded by FTH1), a biomarker of body iron overload, were significantly associated with a high prevalence of hand OA (P-for-trend=0.037).
Conclusion: Our datasets will be valuable as a rich resource and open new possibilities for the research of molecular mechanism, drug development and precise treatment for hand OA. Inflammatory and fibrocartilage chondrocytes are key subpopulations and ferroptosis may be a key pathway in hand OA. Markers of these chondrocyte subpopulations, as well as ferroptosis inhibitors or iron chelators, could be focus of attention in future studies.
.jpg)
H. Li: None; X. Jiang: None; Y. Xiao: None; Y. Zhang: None; W. Zhang: None; M. Doherty: None; J. Nestor: None; C. Li: None; J. Ye: None; T. Sha: None; H. Lyu: None; J. Wei: None; C. Zeng: None; G. Lei: None.
Background/Purpose: Hand osteoarthritis (OA) is a common heterogeneous joint disorder with differences in etiology and pathophysiology from the large weight-bearing knee or hip OA. Its pathophysiological mechanism remains largely unexplored, partially because of limited access to clinical sample tissues and lack of animal models. To date, there is no known cure for hand OA, indicating an urgent need for better understanding of the underlying mechanisms to develop appropriate treatment strategies. Here, we aimed to identify hand joint chondrocyte subpopulations and investigate the molecular mechanism of hand OA by performing single-cell RNA sequencing (scRNA-seq) analysis, and verify the findings in two large independent population-based studies.
Methods: We obtained hand interphalangeal joints from five donors who had destructive forearm injury. Using scRNA-seq analysis, we analyzed the cellular composition and subpopulation-specific gene expression of hand articular cartilage, and then compared these features between cartilages from joints with macroscopically confirmed OA (n=5) and those without OA (n=5). We further conducted: (1) a Mendelian randomization study using data from the UK Biobank to investigate the causal association between the key differentially expressed gene and hand OA; and (2) a cross-sectional study using data collected from a community-based observational study, i.e., the Xiangya OA (XO) Study, to examine the association between a serum biomarker (encoded by the key gene) and hand OA.
Results: Of 105,142 cells we identified 13 subpopulations, including a novel inflammatory chondrocyte subpopulation that specifically expressed genes related to inflammatory and immune response. Fibrocartilage chondrocytes represented a major source of OA-related proteases and exhibited an extensive alteration of gene expression patterns in OA cartilage compared with non-OA cartilage. Both inflammatory chondrocytes and fibrocartilage chondrocytes showed a trend towards increased numbers in hand OA cartilage. In these two subpopulations from OA cartilage, the ferroptosis pathway was enriched, in which the expression of iron overload-related genes, e.g., FTH1, was elevated. These findings are further validated by two independent population-based studies. Among participants (n=332,668) in the UK Biobank, genetic predisposition to higher expression of FTH1 mRNA significantly increased the risk of hand OA (OR=1.07, 95%CI:1.02-1.11). Among participants (n=1,241) from the XO Study, high levels of serum ferritin (encoded by FTH1), a biomarker of body iron overload, were significantly associated with a high prevalence of hand OA (P-for-trend=0.037).
Conclusion: Our datasets will be valuable as a rich resource and open new possibilities for the research of molecular mechanism, drug development and precise treatment for hand OA. Inflammatory and fibrocartilage chondrocytes are key subpopulations and ferroptosis may be a key pathway in hand OA. Markers of these chondrocyte subpopulations, as well as ferroptosis inhibitors or iron chelators, could be focus of attention in future studies.

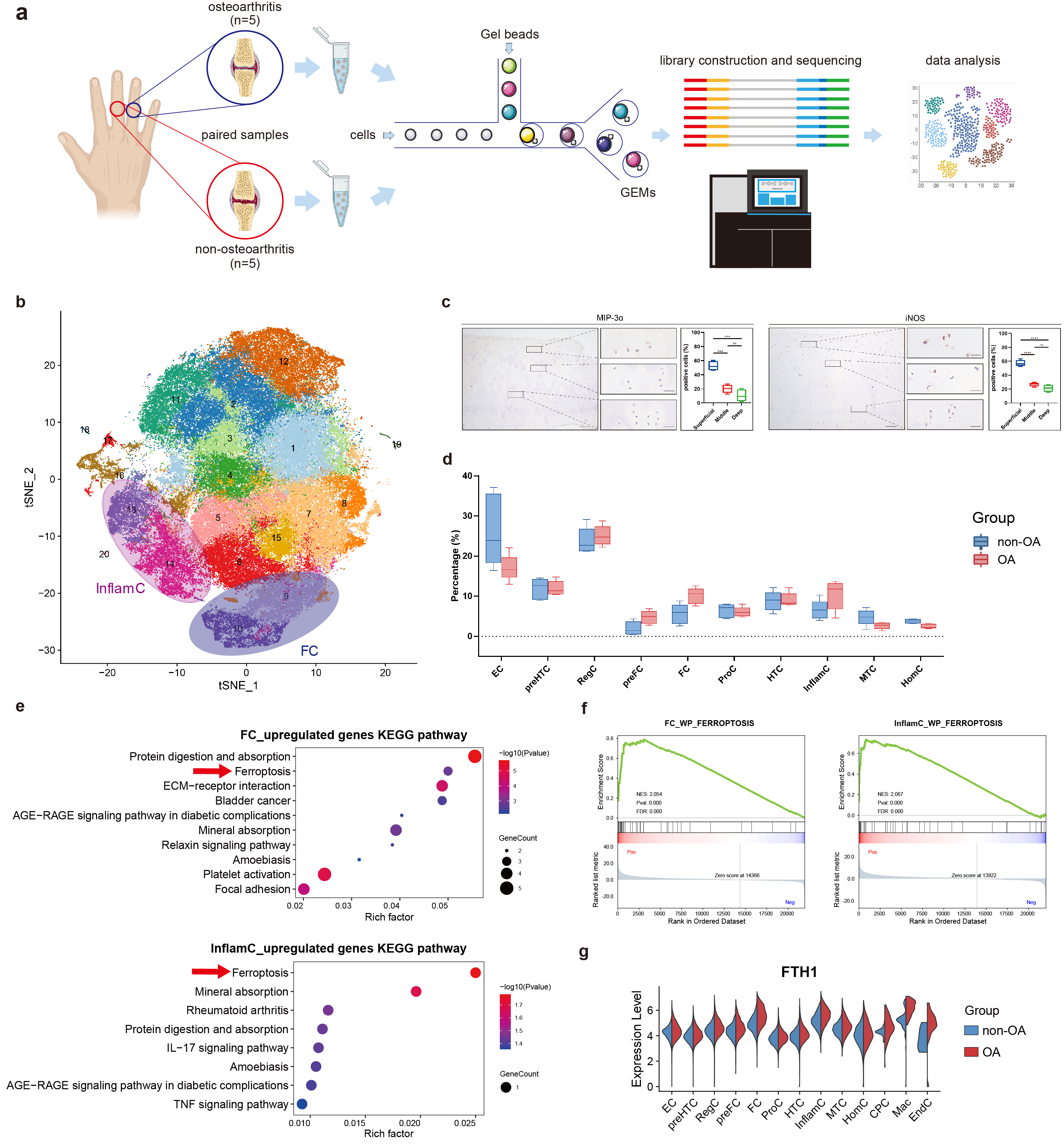
Figure 1. ScRNA-seq analysis of human hand OA and non-OA articular cartilage
(a) Schematic workflow of single-cell RNA sequencing. (b) t-SNE embedding plot for 105,142 cells, derived from paired articular cartilage samples of five donors. (c) Representative immunohistochemistry (IHC) staining for MIP-3α and iNOS in hand articular cartilage tissues (n=4). Scale bar, left, 100 μm; right, 20 μm. ***P<0.001, ****P<0.0001. (d) Differential analysis of subpopulation abundance between osteoarthritic and non-osteoarthritic cartilage. Wilcoxon matched-pairs signed rank test was used for analysis. (e) KEGG enrichment analysis of the upregulated genes of FC and InflamC in hand OA. Arrows indicate ferroptosis enriched in the upregulated genes in osteoarthritic FC and InflamC. (f) GSEA analysis showing extensive enrichment of ferroptosis in the osteoarthritic FC and InflamC. (g) Violin plot showing the differential expression of FTH1 in all subpopulations between OA and non-OA cartilage. OA, osteoarthritis; FC, fibrocartilage chondrocytes; InflamC, inflammatory chondrocytes; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis.
(a) Schematic workflow of single-cell RNA sequencing. (b) t-SNE embedding plot for 105,142 cells, derived from paired articular cartilage samples of five donors. (c) Representative immunohistochemistry (IHC) staining for MIP-3α and iNOS in hand articular cartilage tissues (n=4). Scale bar, left, 100 μm; right, 20 μm. ***P<0.001, ****P<0.0001. (d) Differential analysis of subpopulation abundance between osteoarthritic and non-osteoarthritic cartilage. Wilcoxon matched-pairs signed rank test was used for analysis. (e) KEGG enrichment analysis of the upregulated genes of FC and InflamC in hand OA. Arrows indicate ferroptosis enriched in the upregulated genes in osteoarthritic FC and InflamC. (f) GSEA analysis showing extensive enrichment of ferroptosis in the osteoarthritic FC and InflamC. (g) Violin plot showing the differential expression of FTH1 in all subpopulations between OA and non-OA cartilage. OA, osteoarthritis; FC, fibrocartilage chondrocytes; InflamC, inflammatory chondrocytes; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis.

Figure 2. Mendelian randomization analysis of the causal association between FTH1 mRNA expression and risk of hand OA
.jpg)
Table 1. Association between serum ferritin levels and prevalence of hand OA
H. Li: None; X. Jiang: None; Y. Xiao: None; Y. Zhang: None; W. Zhang: None; M. Doherty: None; J. Nestor: None; C. Li: None; J. Ye: None; T. Sha: None; H. Lyu: None; J. Wei: None; C. Zeng: None; G. Lei: None.



