Abstract Session
Genetics, genomics and proteomics
Session: Abstracts: Genetics, Genomics & Proteomics (0733–0738)
0735: Deep Immunophenotyping Reveals Circulating Activated Lymphocytes in Individuals at Risk for Rheumatoid Arthritis
Sunday, November 12, 2023
2:30 PM - 2:40 PM PT
Location: Room 32A-B
- JI
Jun Inamo, MD, PhD
University of Colorado School of Medicine
Aurora, CO, United StatesDisclosure information not submitted.
Presenting Author(s)
Jun Inamo1, Joshua Keegan2, Alec Griffith2, Tusharkanti Ghosh3, Alice Horisberger2, Kaitlyn Howard2, John Pulford2, Ekaterina Murzin2, Brandon Hancock2, The Accelerating Medicines Partnership SLE/RA4, Marie Feser4, Jill Norri5, Anna Helena Jonsson6, Ye Cao7, William Apruzzese8, S. Louis Bridges9, Vivian Bykerk10, Susan Goodman9, Laura Donlin9, Gary S Firestein11, Harris Perlman12, Joan Bathon13, Laura Hughes14, Darren Tabechian15, Andrew Filer16, Costantino Pitzalis17, Jennifer Anolik15, Larry Moreland18, Joel Guthridge19, Judith James19, Michael Brenner2, Soumya Raychaudhuri6, Jeffrey Sparks20, The Accelerating Medicines Partnership RA/SLE Network21, Michael Holer4, kevin Deane4, James Lederer2, Deepak Rao6 and Fan Zhang22, 1University of Colorado School of Medicine, Aurora, CO, 2Brigham and Women's Hospital and Harvard Medical School, Boston, MA, 3School of Public Health, University of Colorado, Anschutz Medical Campus, Aurora, CO, 4University of Colorado Anschutz Medical Campus, Aurora, CO, 5Colorado School of Public Health, Denver, CO, 6Brigham and Women's Hospital, Boston, MA, 7Brigham and Women's Hospital, Boston, MA, 8Accelerating Medicines Partnership® Program: Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP® RA/SLE) Network, Boston, MA, 9Hospital for Special Surgery, New York, NY, 10Department of Rheumatology, Hospital for Special Surgery, New York, NY, 11Department of Medicine, University of California San Diego, La Jolla, CA, 12Northwestern University, Chicago, IL, 13Columbia University, New York, NY, 14University of Alabama at Birmingham Medicine, Birmingham, AL, 15University of Rochester Medical Center, Rochester, NY, 16University of Birmingham, Birmingham, United Kingdom, 17Queen Mary University of London, London, United Kingdom, 18University of Colorado, Denver, CO, 19Oklahoma Medical Research Foundation, Oklahoma City, OK, 20Division of Rheumatology, Inflammation, and Immunity, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, 21The Accelerating Medicines Partnership RA/SLE Network, 22University of Colorado, Aurora, CO
Background/Purpose: Rheumatoid arthritis (RA) is a systemic autoimmune disease with currently no effective prevention strategies. Single-cell technologies have been used to investigate established RA heterogeneity (1), but it is unknown if the immune populations identified from RA tissues play important roles in blood during the preclinical phase of disease. Thus, identifying pathogenic immune phenotypes in individuals who are at risk for future RA, designated “At-Risk RA”, is crucial to establishing prevention strategies.
Methods: We applied mass cytometry to deeply characterize immunophenotypes in PBMCs (peripheral blood mononuclear cells) from At-Risk individuals based on the presence of antibodies to citrullinated protein antigens (ACPA) and/or first-degree relative (FDR) status (n=52), established RA (n=67), and healthy controls (n=48) (Figure 1). We performed co-varying neighborhood analysis to characterize immunophenotypes in At-Risk individuals and identify phenotypical changes between At-Risk subpopulations accounting for batch effect, age, sex, and inter-individual variation. We further developed an “RA immunophenotype score” for cross-phenotype classification using mixed-effect modeling and logistic regression.
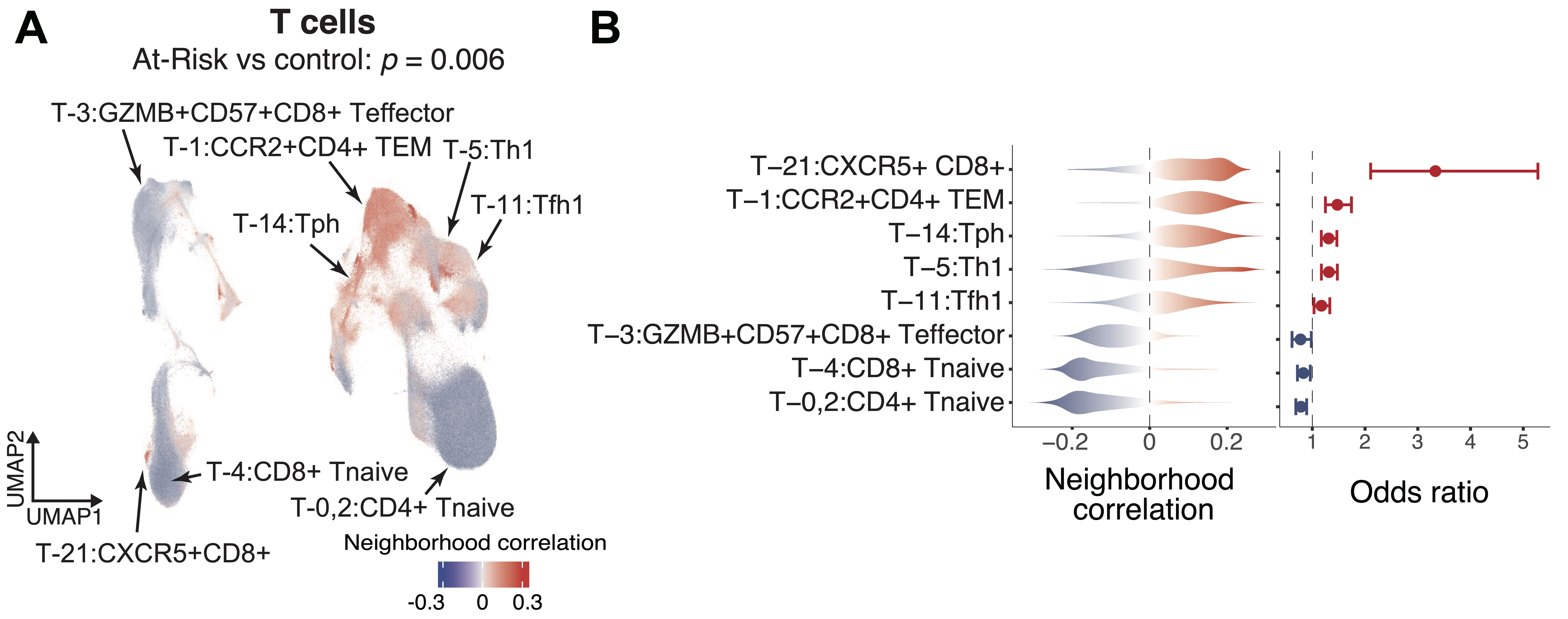
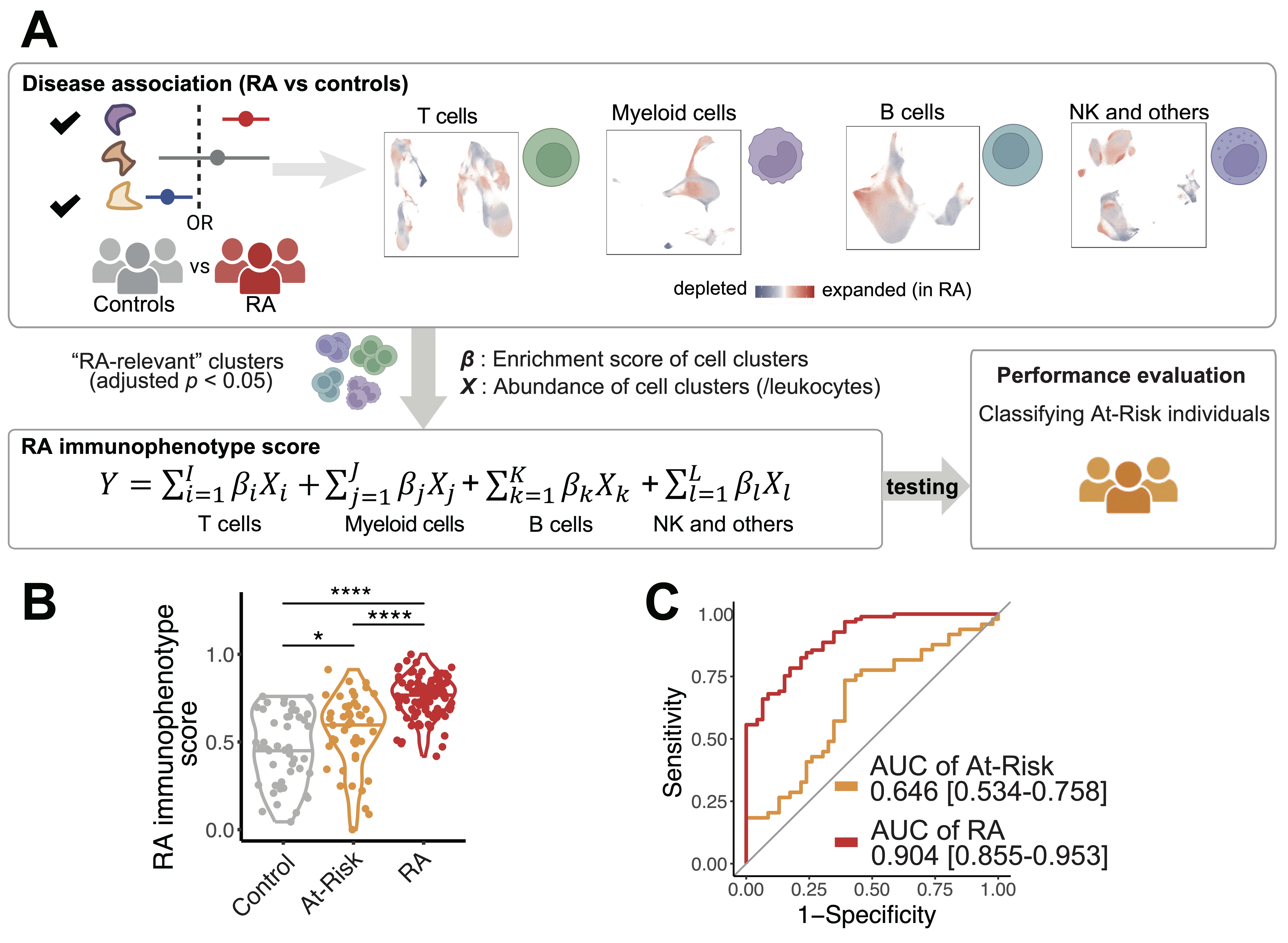
Results: We quantified the immune populations and uncovered significant cell expansions in At-Risk individuals compared with controls (p = 6e-3), including CCR2+CD4+ effector memory T cells (TEM) (OR = 1.47), T peripheral helper cells (Tph) (OR = 1.30), type 1 T helper cells (OR = 1.31), and CXCR5+CD8+ T cells (OR = 3.33) (Figure 2A-B). We further confirmed the expansions of these T cell phenotypes in the At-Risk individuals using our validation cohort with 57 At-Risk and 23 healthy individuals. In addition, we found that CD15+ classical monocytes were especially expanded in ACPA-negative At-Risk individuals who had FDR (p = 1e-3, OR = 1.30), and an activated PAX5low naïve B cell population expanded in ACPA-positive individuals who also had an FDR with RA (p = 9e-3, OR = 1.35). Further, we demonstrated that our “RA immunophenotype score” classification method built based on the degree of enrichment and the abundance of cell states relative to established RA (adjusted p < 0.05) (Figure 3A) is able to significantly distinguish At-Risk individuals from the control (p = 0.039, AUC >0.6) (Figure 3B-C).
Conclusion: We systematically characterized altered circulating immune phenotypes in At-Risk individuals, along with immunophenotypical differences among ACPA+ and FDR At-Risk subpopulations. Our classification model may provide a promising approach for understanding the pathogenesis of preclinical RA with the goal to develop preventive strategies and novel therapeutic targets.
References:
1. Zhang, F. et al. Cellular deconstruction of inflamed synovium defines diverse inflammatory phenotypes in rheumatoid arthritis. bioRxiv (2022) doi: https://doi.org/10.1101/2022.02.25.481990
.jpg)
J. Inamo: None; J. Keegan: None; A. Griffith: None; T. Ghosh: None; A. Horisberger: None; K. Howard: None; J. Pulford: None; E. Murzin: None; B. Hancock: None; T. SLE/RA: None; M. Feser: None; J. Norri: None; A. Jonsson: None; Y. Cao: None; W. Apruzzese: Pfizer, 3; S. Bridges: None; V. Bykerk: AbbVie, 2, Bristol Myers Squibb, 1, 2, 5, Pfizer, 1, 2; S. Goodman: NIH, 5, Novartis, 5; L. Donlin: Bristol-Myers Squibb(BMS), 2, Stryker, 2; G. Firestein: Eli Lilly, 5; H. Perlman: None; J. Bathon: None; L. Hughes: None; D. Tabechian: amgen, 12, share holder; A. Filer: Bristol-Myers Squibb(BMS), 5, GlaxoSmithKlein(GSK), 5, Janssen, 5, Nascient, 5, Sonoma Biotherapeutics, 2; C. Pitzalis: None; J. Anolik: None; L. Moreland: Boehringer-Ingelheim, 12, member of independent Data Safety Monitoring Board, Celltrion, 12, member of independent Data Safety Monitoring Board; J. Guthridge: None; J. James: Bristol-Myers Squibb(BMS), 5, GlaxoSmithKlein(GSK), 2, Novartis, 2, Progentec Biosciences, 5; M. Brenner: 4FO Ventures, 2, GlaxoSmithKlein(GSK), 2, Mestag Therapeutics, 2, 8, Third Rock Ventures, 2; S. Raychaudhuri: AbbVie, 6, Janssen, 1, Mestag, Inc, 2, 8, Pfizer, 1, Sanofi, 1, Sonoma, 1, 8; J. Sparks: AbbVie, 2, Amgen, 2, Boehringer Ingelheim, 2, Bristol-Myers Squibb, 2, 5, Gilead, 2, Inova Diagnostics, 2, Janssen, 2, Optum, 2, Pfizer, 2, ReCor, 2; T. RA/SLE Network: None; M. Holer: None; k. Deane: Bristol-Myers Squibb(BMS), 1, Gilead, 5, Janssen, 5, Werfen, 1, 12, Biomarker kits; J. Lederer: None; D. Rao: AstraZeneca, 2, Bristol-Myers Squibb, 2, 5, GlaxoSmithKlein(GSK), 2, Hifibio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2; F. Zhang: None.
Background/Purpose: Rheumatoid arthritis (RA) is a systemic autoimmune disease with currently no effective prevention strategies. Single-cell technologies have been used to investigate established RA heterogeneity (1), but it is unknown if the immune populations identified from RA tissues play important roles in blood during the preclinical phase of disease. Thus, identifying pathogenic immune phenotypes in individuals who are at risk for future RA, designated “At-Risk RA”, is crucial to establishing prevention strategies.
Methods: We applied mass cytometry to deeply characterize immunophenotypes in PBMCs (peripheral blood mononuclear cells) from At-Risk individuals based on the presence of antibodies to citrullinated protein antigens (ACPA) and/or first-degree relative (FDR) status (n=52), established RA (n=67), and healthy controls (n=48) (Figure 1). We performed co-varying neighborhood analysis to characterize immunophenotypes in At-Risk individuals and identify phenotypical changes between At-Risk subpopulations accounting for batch effect, age, sex, and inter-individual variation. We further developed an “RA immunophenotype score” for cross-phenotype classification using mixed-effect modeling and logistic regression.
Results: We quantified the immune populations and uncovered significant cell expansions in At-Risk individuals compared with controls (p = 6e-3), including CCR2+CD4+ effector memory T cells (TEM) (OR = 1.47), T peripheral helper cells (Tph) (OR = 1.30), type 1 T helper cells (OR = 1.31), and CXCR5+CD8+ T cells (OR = 3.33) (Figure 2A-B). We further confirmed the expansions of these T cell phenotypes in the At-Risk individuals using our validation cohort with 57 At-Risk and 23 healthy individuals. In addition, we found that CD15+ classical monocytes were especially expanded in ACPA-negative At-Risk individuals who had FDR (p = 1e-3, OR = 1.30), and an activated PAX5low naïve B cell population expanded in ACPA-positive individuals who also had an FDR with RA (p = 9e-3, OR = 1.35). Further, we demonstrated that our “RA immunophenotype score” classification method built based on the degree of enrichment and the abundance of cell states relative to established RA (adjusted p < 0.05) (Figure 3A) is able to significantly distinguish At-Risk individuals from the control (p = 0.039, AUC >0.6) (Figure 3B-C).
Conclusion: We systematically characterized altered circulating immune phenotypes in At-Risk individuals, along with immunophenotypical differences among ACPA+ and FDR At-Risk subpopulations. Our classification model may provide a promising approach for understanding the pathogenesis of preclinical RA with the goal to develop preventive strategies and novel therapeutic targets.
References:
1. Zhang, F. et al. Cellular deconstruction of inflamed synovium defines diverse inflammatory phenotypes in rheumatoid arthritis. bioRxiv (2022) doi: https://doi.org/10.1101/2022.02.25.481990
.jpg)
Fig.1: Overview of mass cytometry analytical strategy, clustering, and classifications for At-Risk RA and established RA individuals.

Fig.2: Identifications of specific T cell populations that were associated with At-Risk. A. Cells in UMAP are colored in red (expansion) or blue (depletion) and p-value is shown as well. B. Distributions of cell neighborhood correlations and odds ratio. Error bars represent 95% confidence intervals.

Fig.3: Classifications for At-Risk individuals and established RA individuals. A. RA immunophenotype score utilizing RA-specific cell type abundances to quantify and distinguish At-Risk individuals from control. For each cell type, all p-values from the covarying neighborhood analysis test were p = 1e-3. We incorporated clusters that are significantly associated with RA (adjusted p < 0.05) to model the RA immunophenotype score. We calculated RA immunophenotype score based on cell type abundances multiplied by corresponding major cell type proportions and enrichment scores for each cell type, B. Distribution of RA immunophenotype score across individual samples from RA, At-Risk, and controls; **** p < 0.0001, * p < 0.05, C. Receiver operating characteristic (ROC) analysis to evaluate the classification performance of RA immunophenotype score in distinguishing At-Risk from control. Areas under the curve (AUC) with 95% confidence intervals were described. All the analyses are adjusted for age and sex.
J. Inamo: None; J. Keegan: None; A. Griffith: None; T. Ghosh: None; A. Horisberger: None; K. Howard: None; J. Pulford: None; E. Murzin: None; B. Hancock: None; T. SLE/RA: None; M. Feser: None; J. Norri: None; A. Jonsson: None; Y. Cao: None; W. Apruzzese: Pfizer, 3; S. Bridges: None; V. Bykerk: AbbVie, 2, Bristol Myers Squibb, 1, 2, 5, Pfizer, 1, 2; S. Goodman: NIH, 5, Novartis, 5; L. Donlin: Bristol-Myers Squibb(BMS), 2, Stryker, 2; G. Firestein: Eli Lilly, 5; H. Perlman: None; J. Bathon: None; L. Hughes: None; D. Tabechian: amgen, 12, share holder; A. Filer: Bristol-Myers Squibb(BMS), 5, GlaxoSmithKlein(GSK), 5, Janssen, 5, Nascient, 5, Sonoma Biotherapeutics, 2; C. Pitzalis: None; J. Anolik: None; L. Moreland: Boehringer-Ingelheim, 12, member of independent Data Safety Monitoring Board, Celltrion, 12, member of independent Data Safety Monitoring Board; J. Guthridge: None; J. James: Bristol-Myers Squibb(BMS), 5, GlaxoSmithKlein(GSK), 2, Novartis, 2, Progentec Biosciences, 5; M. Brenner: 4FO Ventures, 2, GlaxoSmithKlein(GSK), 2, Mestag Therapeutics, 2, 8, Third Rock Ventures, 2; S. Raychaudhuri: AbbVie, 6, Janssen, 1, Mestag, Inc, 2, 8, Pfizer, 1, Sanofi, 1, Sonoma, 1, 8; J. Sparks: AbbVie, 2, Amgen, 2, Boehringer Ingelheim, 2, Bristol-Myers Squibb, 2, 5, Gilead, 2, Inova Diagnostics, 2, Janssen, 2, Optum, 2, Pfizer, 2, ReCor, 2; T. RA/SLE Network: None; M. Holer: None; k. Deane: Bristol-Myers Squibb(BMS), 1, Gilead, 5, Janssen, 5, Werfen, 1, 12, Biomarker kits; J. Lederer: None; D. Rao: AstraZeneca, 2, Bristol-Myers Squibb, 2, 5, GlaxoSmithKlein(GSK), 2, Hifibio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2; F. Zhang: None.



