Late-Breaking Abstract Session
Sjögren’s syndrome
Session: Late-Breaking Abstracts (L15–L20)
L18: Immune-metabolic Heterogeneity and Clinical Implications in Primary Sjogren's Syndrome Revealed by Molecular Classification of Salivary Glands
Wednesday, November 15, 2023
8:15 AM - 8:30 AM PT
Location: Room 6A-B
- HX
Huji Xu, n/a
Department of Rheumatology and Immunology, Changzheng Hospital, Naval Medical University
Shanghai, ChinaDisclosure information not submitted.
Presenting Author(s)
Xiaobing Wang1, Jing Luo2, Senhong Ying3, Jingwei Hong4, Hui Cheng4, Ping Wang4, Yanran He5, Wenjing Ye6, Xiaofang Zhu7, Chengwei Zhu8, Langxiong Yang2, Zhongshan Li9, Suxian Lin10, Dan Chen7, Xin Wu11, Zhengwei Xie12, Jinyu Wu13 and Huji Xu1, 1Department of Rheumatology and Immunology, Changzheng Hospital, Naval Medical University, Shanghai, China, 2School of Medicine, Tsinghua University, Beijing, China, 3Precision Medicine Center, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China, 4Department of Rheumatology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China, 5Committee on Cancer Biology, University of Chicago, Chicago, IL, 6Division of Rheumatology, Huashan Hospital, Fudan University, Shanghai, China, 7Rheumatology Department, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China, 8Department of Ultrasonography, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China, 9Wenzhou Medical University, Wenzhou, China, 10Rheumatology Department, Wenzhou People's Hospital, Wenzhou, China, 11Department of Rheumatology and Immunology,Changzheng Hospital, Naval Medical University, Shanghai, China, 12Peking University International Cancer Institute, Health Science Center, Peking University, Beijing, China, 13Institute of Genomic Medicine, Key Laboratory of Laboratory Medicine, Ministry of Education, Wenzhou Medical University, Wenzhou, China
Background/Purpose: Primary Sjögren's syndrome (pSS) is a complex autoimmune disease with significant heterogeneity. Our study aimed to clarify the etiology and molecular variation of the target organ, salivary glands (SGs), in pSS by stratifying them into distinguishable subgroups, which could inform treatment choices in pSS.
Methods: Large-scale transcriptomic profiling of SGs was conducted on 396 pSS, 87 non-pSS, and 44 early-pSS individuals from a multicenter consecutive Chinese cohort. Unsupervised clustering methods were used for molecular classification and integrated analyses were performed to elucidate comprehensive clinical and biological features, immune-metabolic and genetic characteristics. Lymphoma risk and treatment response for pSS subgroups was also predicted for pSS subtypes.
Results: Three distinct subtypes were identified both in established and early-pSS, including "pauci-immune/C1", "cold-immune/C2", and "hot-immune/C3". Pauci-immune/C1 exhibited a similar biologic pattern to non-pSS. The cold-immune/C2 displayed dramatic activation of classical adaptive immune and depressed metabolism, along with mitochondrial dysfunction and cGAS-STING-NFkB signaling overactivity. “Hot-immune/C3” had an innate immune inclination and predominantly active metabolism processes, especially for oxidative phosphorylation (OXPHOS). Among the three subtypes, C2 had a higher lymphoma risk with the highest immune cell and endothelial cell infiltration in SGs. Based on the distinct features of each group, we matched them with approved or exploratory treatment options the patient most likely would benefit from, including conventional medicine, JAK inhibitors, and biologics.
Conclusion: Overall, our findings provide a comprehensive and in-depth molecular landscape of SGs in pSS, shedding light on the disease heterogeneity and promoting the development of precise clinical intervention strategies for pSS patients.
Immune-metabolic Heterogeneity and Clinical Implications in Primary Sjogren's Syndrome Revealed by Molecular Classification of Salivary Glands
X. Wang: None; J. Luo: None; S. Ying: None; J. Hong: None; H. Cheng: None; P. Wang: None; Y. He: None; W. Ye: None; X. Zhu: None; C. Zhu: None; L. Yang: None; Z. Li: None; S. Lin: None; D. Chen: None; X. Wu: None; Z. Xie: None; J. Wu: None; H. Xu: None.
Background/Purpose: Primary Sjögren's syndrome (pSS) is a complex autoimmune disease with significant heterogeneity. Our study aimed to clarify the etiology and molecular variation of the target organ, salivary glands (SGs), in pSS by stratifying them into distinguishable subgroups, which could inform treatment choices in pSS.
Methods: Large-scale transcriptomic profiling of SGs was conducted on 396 pSS, 87 non-pSS, and 44 early-pSS individuals from a multicenter consecutive Chinese cohort. Unsupervised clustering methods were used for molecular classification and integrated analyses were performed to elucidate comprehensive clinical and biological features, immune-metabolic and genetic characteristics. Lymphoma risk and treatment response for pSS subgroups was also predicted for pSS subtypes.
Results: Three distinct subtypes were identified both in established and early-pSS, including "pauci-immune/C1", "cold-immune/C2", and "hot-immune/C3". Pauci-immune/C1 exhibited a similar biologic pattern to non-pSS. The cold-immune/C2 displayed dramatic activation of classical adaptive immune and depressed metabolism, along with mitochondrial dysfunction and cGAS-STING-NFkB signaling overactivity. “Hot-immune/C3” had an innate immune inclination and predominantly active metabolism processes, especially for oxidative phosphorylation (OXPHOS). Among the three subtypes, C2 had a higher lymphoma risk with the highest immune cell and endothelial cell infiltration in SGs. Based on the distinct features of each group, we matched them with approved or exploratory treatment options the patient most likely would benefit from, including conventional medicine, JAK inhibitors, and biologics.
Conclusion: Overall, our findings provide a comprehensive and in-depth molecular landscape of SGs in pSS, shedding light on the disease heterogeneity and promoting the development of precise clinical intervention strategies for pSS patients.

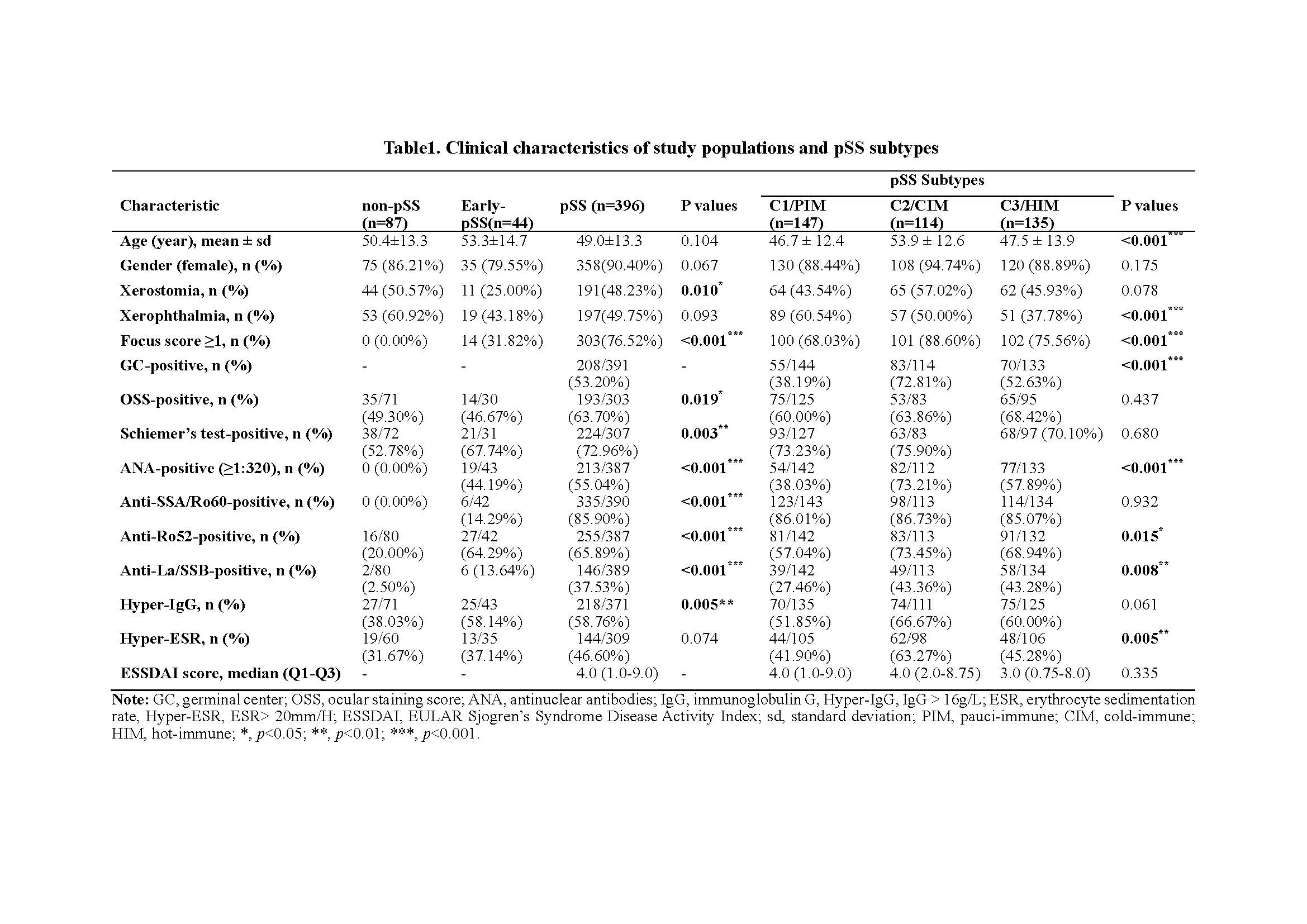
Table1. Clinical characteristics of study populations and pSS subtypes

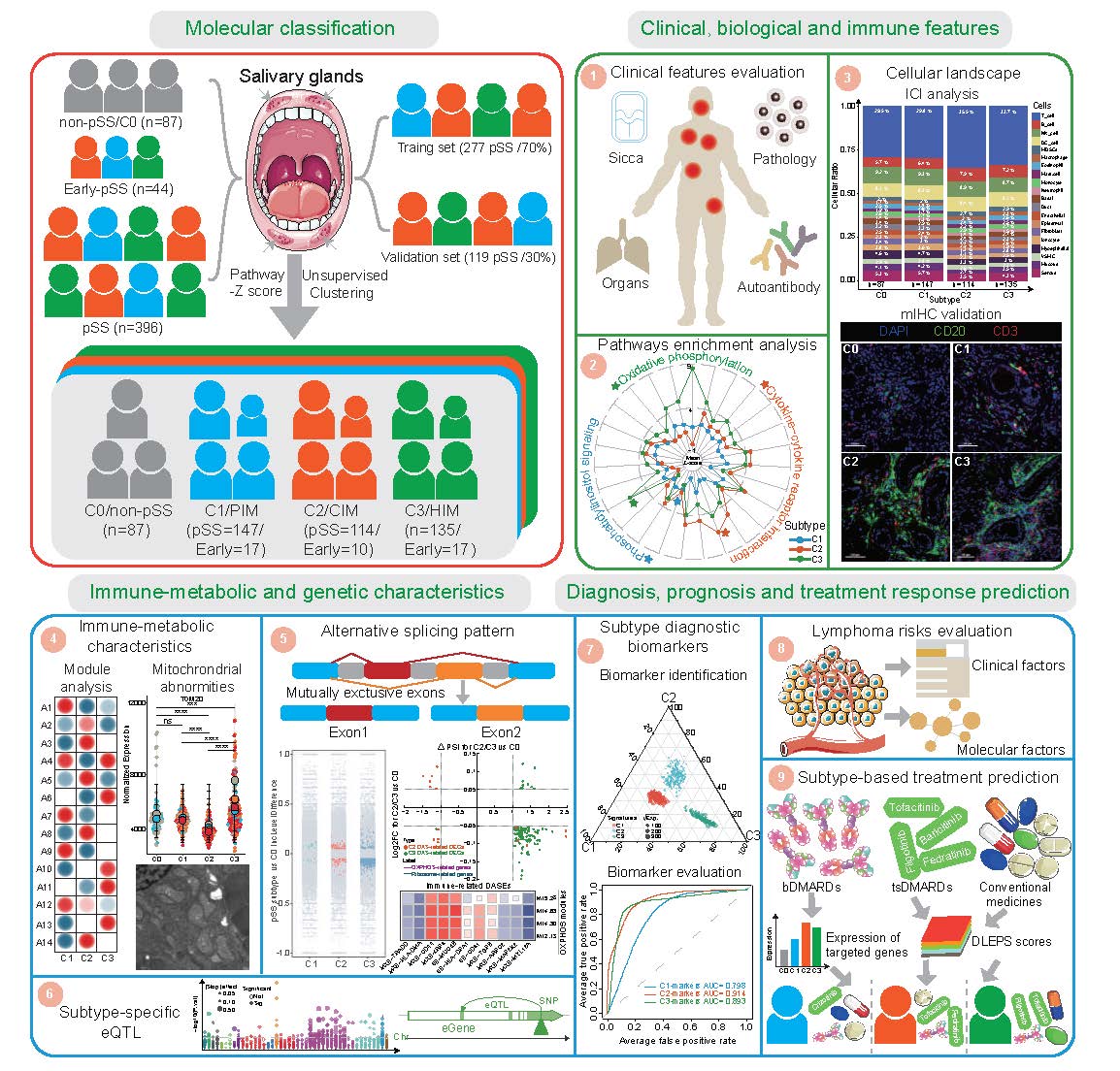
Overview of this study
We recruited 527 participants based on respective criteria, including 87 non-pSS (C0), 44 early-pSS, and 396 pSS. Using an unsupervised clustering approach on the transcriptomic data of SGs, we identified three distinct molecular subtypes in pSS and early-pSS. We then conducted extensive downstream analysis on their clinical and biological and immune features, immune-metabolic and genetic characteristics, and disease diagnosis, prognosis, and treatment response prediction.
pSS, primary Sjogren's syndrome; PIM, pauci-immune; CIM, cold-immune; HIM, heat-immune; ICI, immune cell infiltration; mIHC, multiplex immunohistochemistry; eQTL, expression quantitative trait loci; bDMARDs, biologic disease-modifying anti-rheumatic drugs; tsDMARDs, targeted synthesis disease-modifying anti-rheumatic drugs.
We recruited 527 participants based on respective criteria, including 87 non-pSS (C0), 44 early-pSS, and 396 pSS. Using an unsupervised clustering approach on the transcriptomic data of SGs, we identified three distinct molecular subtypes in pSS and early-pSS. We then conducted extensive downstream analysis on their clinical and biological and immune features, immune-metabolic and genetic characteristics, and disease diagnosis, prognosis, and treatment response prediction.
pSS, primary Sjogren's syndrome; PIM, pauci-immune; CIM, cold-immune; HIM, heat-immune; ICI, immune cell infiltration; mIHC, multiplex immunohistochemistry; eQTL, expression quantitative trait loci; bDMARDs, biologic disease-modifying anti-rheumatic drugs; tsDMARDs, targeted synthesis disease-modifying anti-rheumatic drugs.

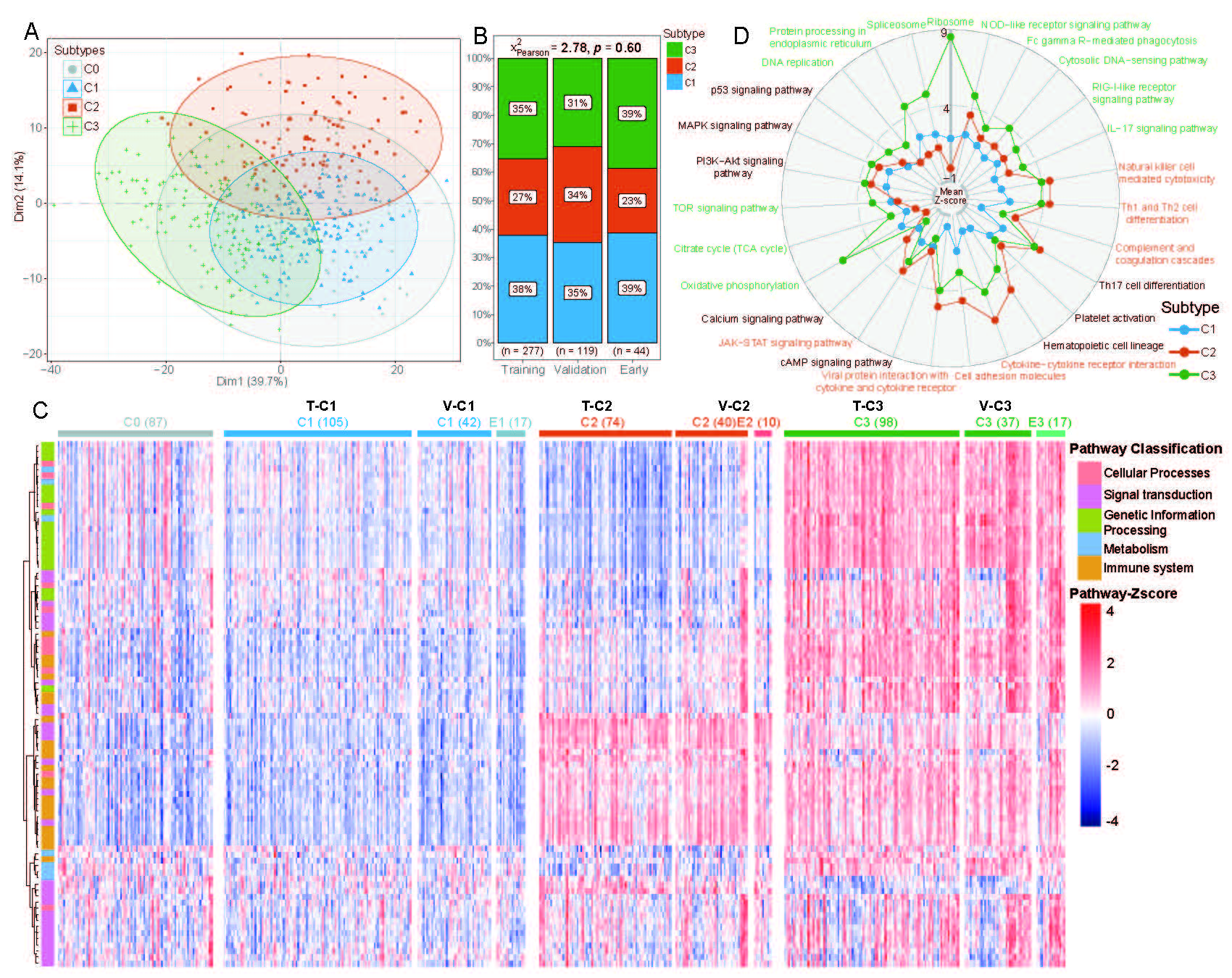
Molecular classifications and biological features of pSS subtypes
(A) A PCA plot to show 3 groups of pSS patients (C1, C2, C3) by hierarchical pathway-based clustering. Non-pSS cases were overlaid using the same algorithm (C0, grey dots).
(B) The scale map showing the distribution of 3 pSS subgroups in the Training set, Validation set and early-pSS cohorts without significant difference (Chi-Square test, p =0.60).
(C) A heatmap generated from transcriptomic information via hierarchical pathway-based clustering based on z-score levels, including pSS Training set: 277/70%, pSS Validation set: 119/30%, 44 early-pSS patients, and 87 non-pSS cohorts, illustrating the distribution of five categories of pathways across the ten subtypes (non-pSS/C0 (87), T-C1(105), V-C1(42), E1(17), T-C2(74), V-C2 (40), E2(10), T-C3(98), V-C3(37) and E3(17)). T-C represents the clustering pSS subtypes in the Training set; V-C represents the clustering pSS subtypesthe in the Validation set; E represents the clustering early-pSS subtypes.
(D) Representative significant pathways for each cluster. A radar plot showing the mean pathway z-scores in three pSS subtypes; C1 (blue), C2 (red), and C3 (green).
(A) A PCA plot to show 3 groups of pSS patients (C1, C2, C3) by hierarchical pathway-based clustering. Non-pSS cases were overlaid using the same algorithm (C0, grey dots).
(B) The scale map showing the distribution of 3 pSS subgroups in the Training set, Validation set and early-pSS cohorts without significant difference (Chi-Square test, p =0.60).
(C) A heatmap generated from transcriptomic information via hierarchical pathway-based clustering based on z-score levels, including pSS Training set: 277/70%, pSS Validation set: 119/30%, 44 early-pSS patients, and 87 non-pSS cohorts, illustrating the distribution of five categories of pathways across the ten subtypes (non-pSS/C0 (87), T-C1(105), V-C1(42), E1(17), T-C2(74), V-C2 (40), E2(10), T-C3(98), V-C3(37) and E3(17)). T-C represents the clustering pSS subtypes in the Training set; V-C represents the clustering pSS subtypesthe in the Validation set; E represents the clustering early-pSS subtypes.
(D) Representative significant pathways for each cluster. A radar plot showing the mean pathway z-scores in three pSS subtypes; C1 (blue), C2 (red), and C3 (green).
Immune-metabolic Heterogeneity and Clinical Implications in Primary Sjogren's Syndrome Revealed by Molecular Classification of Salivary Glands
X. Wang: None; J. Luo: None; S. Ying: None; J. Hong: None; H. Cheng: None; P. Wang: None; Y. He: None; W. Ye: None; X. Zhu: None; C. Zhu: None; L. Yang: None; Z. Li: None; S. Lin: None; D. Chen: None; X. Wu: None; Z. Xie: None; J. Wu: None; H. Xu: None.




