Poster Session A
Epidemiology, health policy and outcomes
Session: (0117–0144) Epidemiology & Public Health Poster I
0132: Feasibility of a Real-Time Risk Model to Identify Antinuclear Antibody Positive Individuals at Risk for Systemic Autoimmune Disease
Sunday, November 12, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
.png)
April Barnado, MD, MSc
Vanderbilt University Medical Center
Nashville, TN, United StatesDisclosure(s): No financial relationships with ineligible companies to disclose
Abstract Poster Presenter(s)
April Barnado, Ryan Moore, Hank Domenico, Sarah Green, Alex Camai, Ashley Suh, Bryan Han, Katherine Walker, Audrey Anderson, Lannawill Caruth, Anish Katta, Allison McCoy and Daniel W. Byrne, Vanderbilt University Medical Center, Nashville, TN
Background/Purpose: Positive antinuclear antibodies (ANAs) cause diagnostic dilemmas for clinicians across multiple specialties. We previously developed and validated a risk model using a de-identified electronic health record (EHR) database to distinguish individuals with positive ANAs who develop systemic autoimmune diseases from individuals who do not. We assessed the feasibility of deploying the risk model in the EHR. We also examined how the model performed in individuals with different systemic autoimmune diseases.
Methods: This logistic regression model contains the following predictors: age at time of first positive ANA, sex, ANA titer, platelet count, billing codes, and presence of another autoantibody (i.e. dsDNA, SSA). The most important variable in the model was presence of another autoantibody. We applied our risk model to data extracted from our EHR-provided data warehouse (Epic Clarity) to assess feasibility of deploying the model in real-time. We calculated risk probabilities for individuals with positive ANAs from 2017-2021. This time period captured an updated ANA titer reporting to the most current data available. Additionally, we combined training (n = 1030) and validation (n = 449) sets from the de-identified EHR. We compared risk scores for individuals in each of the autoimmune disease categories and in individuals without autoimmune diseases. Risk scores are reported as medians with interquartile ranges (IQR) and compared using the Mann-Whitney U test.
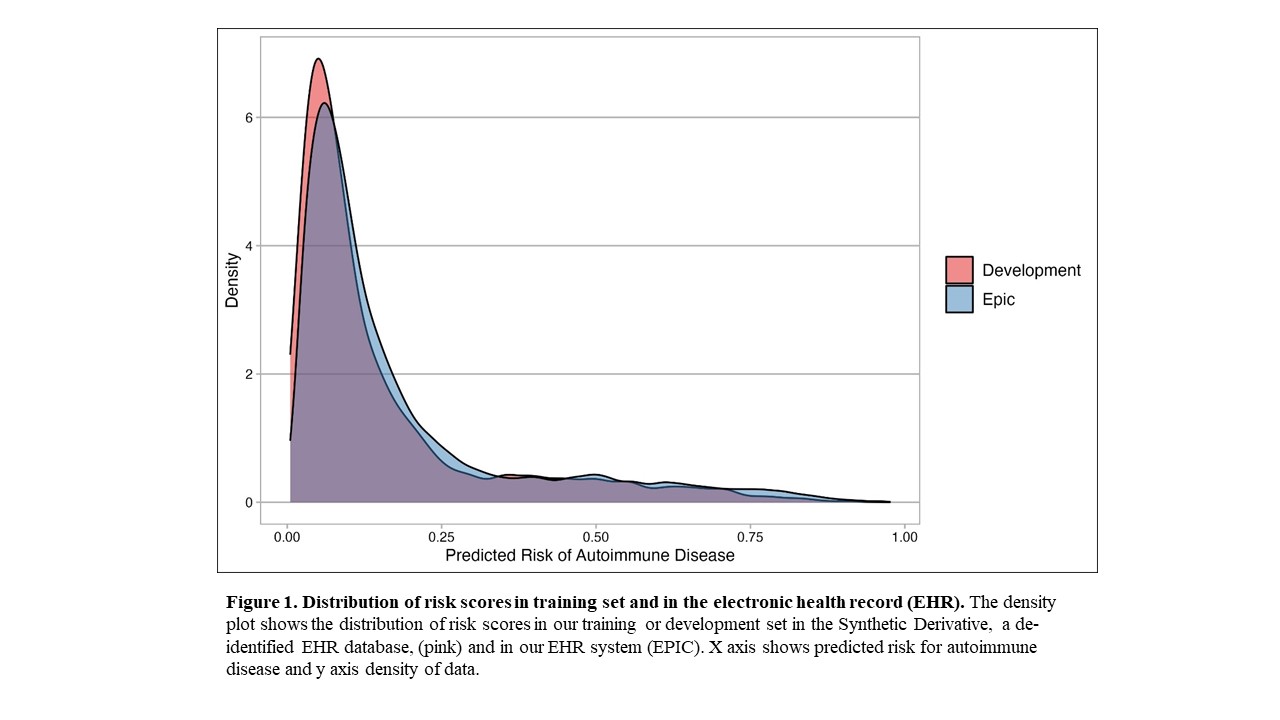
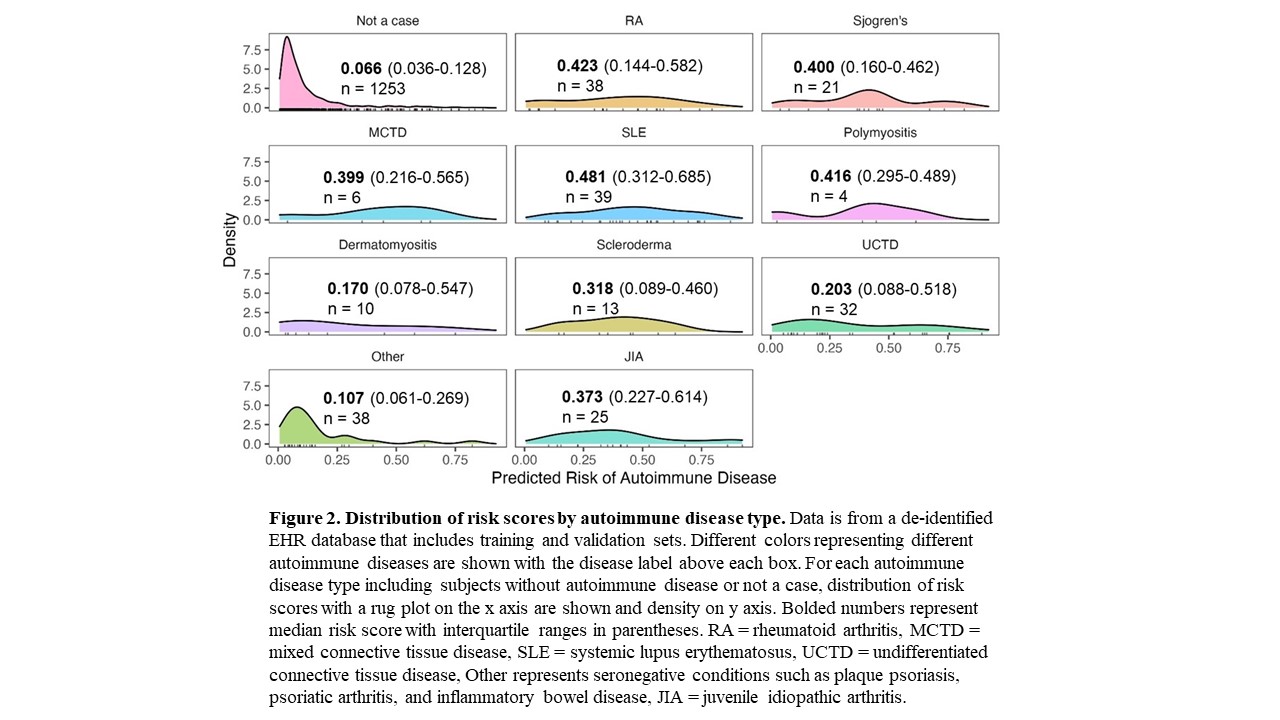
Results: We assessed the risk model using Epic EHR data for all individuals with a positive ANA from 2017-2021 (n = 22,234). We observed a similar distribution of risk scores in Epic compared to our model training set that used a de-identified EHR database (Figure 1). Risk scores for positive ANA individuals with and without autoimmune diseases from the combined training and validation sets from the de-identified EHR are shown in Figure 2. Individuals with systemic lupus erythematosus (SLE) had the highest risk scores with a median of 0.481 and IQR of 0.312-0.685 followed by Rheumatoid Arthritis with 0.423 (0.144-0.582). Individuals labeled as other, with predominantly seronegative conditions, had the lowest median risk score of 0.107 (0.061-0.269). Seronegative conditions included plaque psoriasis, psoriatic arthritis, and inflammatory bowel disease. Individuals with seropositive diseases had a higher risk score compared to individuals with seronegative diseases (p < 0.001). For individuals without autoimmune diseases, when available alternative diagnoses were documented by rheumatologists, the most frequent diagnoses were fibromyalgia, osteoarthritis, and gout.
Conclusion: To our best knowledge, a risk model that focuses on individuals with positive ANAs and predicts risk for multiple systemic autoimmune diseases does not currently exist. We demonstrate a validated risk model that uses readily available EHR data and can be successfully deployed in the EHR. For future studies, we will assess our risk model prospectively in real-time in the EHR and its impact on time to diagnosis and treatment for autoimmune diseases.
A. Barnado: None; R. Moore: None; H. Domenico: None; S. Green: None; A. Camai: None; A. Suh: None; B. Han: None; K. Walker: None; A. Anderson: None; L. Caruth: None; A. Katta: None; A. McCoy: None; D. Byrne: None.
Background/Purpose: Positive antinuclear antibodies (ANAs) cause diagnostic dilemmas for clinicians across multiple specialties. We previously developed and validated a risk model using a de-identified electronic health record (EHR) database to distinguish individuals with positive ANAs who develop systemic autoimmune diseases from individuals who do not. We assessed the feasibility of deploying the risk model in the EHR. We also examined how the model performed in individuals with different systemic autoimmune diseases.
Methods: This logistic regression model contains the following predictors: age at time of first positive ANA, sex, ANA titer, platelet count, billing codes, and presence of another autoantibody (i.e. dsDNA, SSA). The most important variable in the model was presence of another autoantibody. We applied our risk model to data extracted from our EHR-provided data warehouse (Epic Clarity) to assess feasibility of deploying the model in real-time. We calculated risk probabilities for individuals with positive ANAs from 2017-2021. This time period captured an updated ANA titer reporting to the most current data available. Additionally, we combined training (n = 1030) and validation (n = 449) sets from the de-identified EHR. We compared risk scores for individuals in each of the autoimmune disease categories and in individuals without autoimmune diseases. Risk scores are reported as medians with interquartile ranges (IQR) and compared using the Mann-Whitney U test.
Results: We assessed the risk model using Epic EHR data for all individuals with a positive ANA from 2017-2021 (n = 22,234). We observed a similar distribution of risk scores in Epic compared to our model training set that used a de-identified EHR database (Figure 1). Risk scores for positive ANA individuals with and without autoimmune diseases from the combined training and validation sets from the de-identified EHR are shown in Figure 2. Individuals with systemic lupus erythematosus (SLE) had the highest risk scores with a median of 0.481 and IQR of 0.312-0.685 followed by Rheumatoid Arthritis with 0.423 (0.144-0.582). Individuals labeled as other, with predominantly seronegative conditions, had the lowest median risk score of 0.107 (0.061-0.269). Seronegative conditions included plaque psoriasis, psoriatic arthritis, and inflammatory bowel disease. Individuals with seropositive diseases had a higher risk score compared to individuals with seronegative diseases (p < 0.001). For individuals without autoimmune diseases, when available alternative diagnoses were documented by rheumatologists, the most frequent diagnoses were fibromyalgia, osteoarthritis, and gout.
Conclusion: To our best knowledge, a risk model that focuses on individuals with positive ANAs and predicts risk for multiple systemic autoimmune diseases does not currently exist. We demonstrate a validated risk model that uses readily available EHR data and can be successfully deployed in the EHR. For future studies, we will assess our risk model prospectively in real-time in the EHR and its impact on time to diagnosis and treatment for autoimmune diseases.


A. Barnado: None; R. Moore: None; H. Domenico: None; S. Green: None; A. Camai: None; A. Suh: None; B. Han: None; K. Walker: None; A. Anderson: None; L. Caruth: None; A. Katta: None; A. McCoy: None; D. Byrne: None.



