Poster Session A
Immunobiology
Session: (0040–0064) Innate Immunity Poster
0047: Identification of Specific Monocyte Epigenetic Signatures in Sarcoidosis and Tuberculosis
Sunday, November 12, 2023
9:00 AM - 11:00 AM PT
Location: Poster Hall
- AM
Abstract Poster Presenter(s)
Marie Robert1, Nader Yatim2, Arthur Mageau3, Nicolas Charles4, Tiphaine Goulenok2, Tom Dott1, Violaine Saint Andre1, Darragh Duffy1 and Karim Sacre3, 1Institut Pasteur Paris, Paris, France, 2Assistance Publique Hopitaux de Paris, Paris, France, 3Université Paris Cité, Paris, France, 4INSERM, Paris, France
Background/Purpose: Sarcoidosis is an inflammatory disease characterized by granuloma tissue lesions that most commonly affect the lungs, but can target any other organ. While the cause of sarcoidosis and the mechanisms underlying granuloma formation and progression are still unknown, the disease shares many similarities with tuberculosis, including histological characteristics, organ tropism and clinical presentation. We hypothesized that circulating monocytes from patients with sarcoidosis or tuberculosis may retain a specific epigenetic signature, distinguishing from healthy controls and each disease.
Methods: To test this, we performed genome-wide epigenetic profiling of circulating monocytes from newly diagnosed patients with tuberculosis or sarcoidosis using the CUT&Tag technology, and assessed the acetylation of lysine 27 of histone 3 (H3K27Ac) (the most appropriate histone mark to identify enhancers of expressed genes).
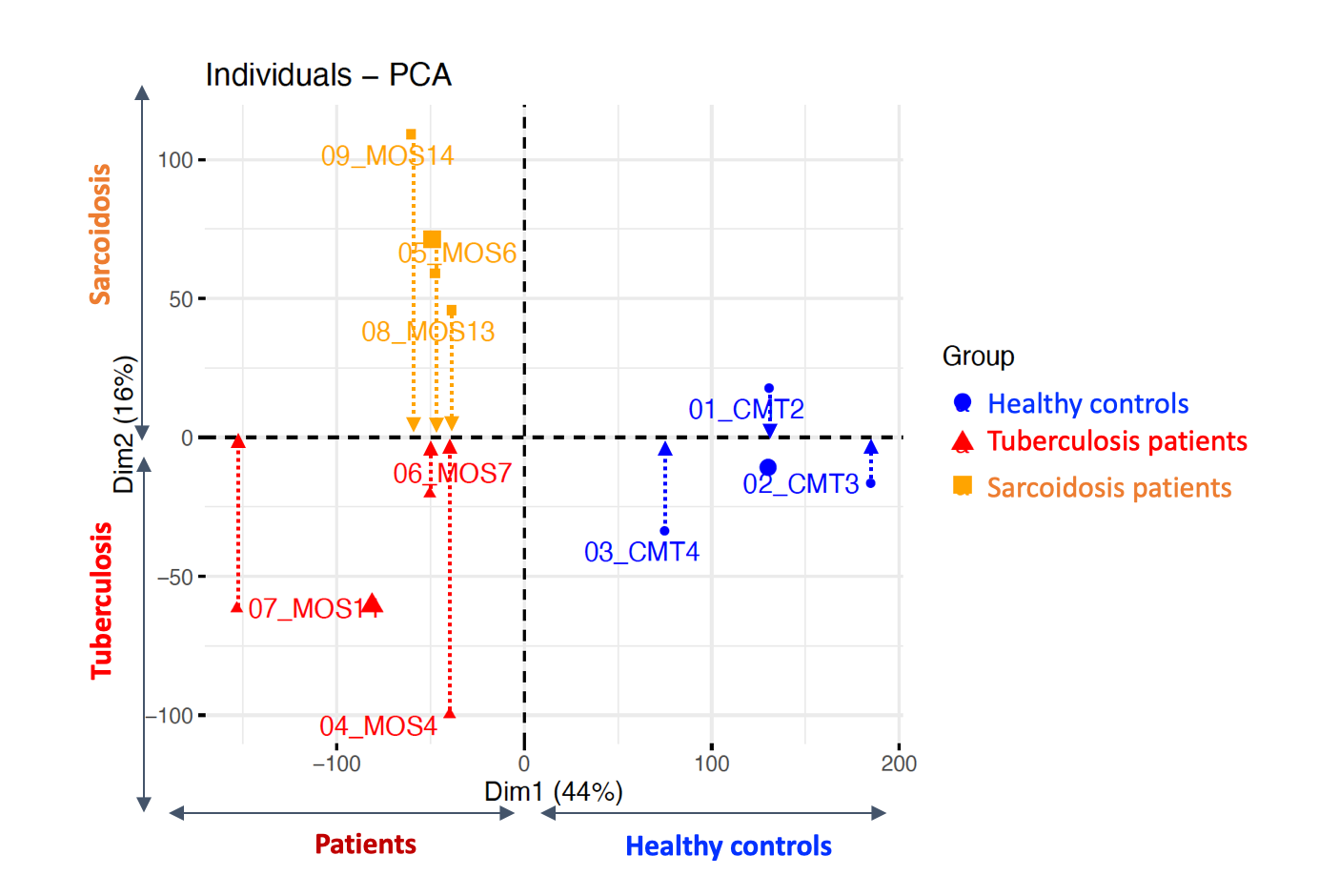
Results: Using unbiased principal component analysis (PCA) of genome-wide histone H3K27Ac profiles we identified genomic regions that specifically separated sarcoidosis patients, from tuberculosis patients and healthy donors (Figure 1). There were 648 H3K27Ac enriched regions of the genome in sarcoidosis as compared to tuberculosis, while 203 regions were H3K27Ac depleted (log2FC > 0.3 or < 0.3, adj-p < 0.1). Interestingly, the genes associated with these regions can discriminate sarcoidosis from tuberculosis patients.
Conclusion: Our findings provide evidence that there are distinct monocyte epigenetic signatures associated with sarcoidosis and tuberculosis, and raise new and challenging perspectives on the role played by trained immunity in inflammatory diseases. Ongoing work will extend this analysis and integrate with additional immune phenotyping and functional assays for a more comprehensive understanding of sarcoidosis.
M. Robert: None; N. Yatim: Hi-Bio, 2; A. Mageau: None; N. Charles: argenx, 2, 5, Onward Therapeutics, 2; T. Goulenok: None; T. Dott: None; V. Saint Andre: None; D. Duffy: Roche, 5, Sanofi, 5; K. Sacre: None.
Background/Purpose: Sarcoidosis is an inflammatory disease characterized by granuloma tissue lesions that most commonly affect the lungs, but can target any other organ. While the cause of sarcoidosis and the mechanisms underlying granuloma formation and progression are still unknown, the disease shares many similarities with tuberculosis, including histological characteristics, organ tropism and clinical presentation. We hypothesized that circulating monocytes from patients with sarcoidosis or tuberculosis may retain a specific epigenetic signature, distinguishing from healthy controls and each disease.
Methods: To test this, we performed genome-wide epigenetic profiling of circulating monocytes from newly diagnosed patients with tuberculosis or sarcoidosis using the CUT&Tag technology, and assessed the acetylation of lysine 27 of histone 3 (H3K27Ac) (the most appropriate histone mark to identify enhancers of expressed genes).
Results: Using unbiased principal component analysis (PCA) of genome-wide histone H3K27Ac profiles we identified genomic regions that specifically separated sarcoidosis patients, from tuberculosis patients and healthy donors (Figure 1). There were 648 H3K27Ac enriched regions of the genome in sarcoidosis as compared to tuberculosis, while 203 regions were H3K27Ac depleted (log2FC > 0.3 or < 0.3, adj-p < 0.1). Interestingly, the genes associated with these regions can discriminate sarcoidosis from tuberculosis patients.
Conclusion: Our findings provide evidence that there are distinct monocyte epigenetic signatures associated with sarcoidosis and tuberculosis, and raise new and challenging perspectives on the role played by trained immunity in inflammatory diseases. Ongoing work will extend this analysis and integrate with additional immune phenotyping and functional assays for a more comprehensive understanding of sarcoidosis.

Figure 1. Unbiased principal component analysis (PCA) of genome-wide histone H3K27Ac profiles using CUT&Tag technology.
M. Robert: None; N. Yatim: Hi-Bio, 2; A. Mageau: None; N. Charles: argenx, 2, 5, Onward Therapeutics, 2; T. Goulenok: None; T. Dott: None; V. Saint Andre: None; D. Duffy: Roche, 5, Sanofi, 5; K. Sacre: None.



