Abstract Session
Genetics, genomics and proteomics
Session: Abstracts: Genetics, Genomics & Proteomics (0733–0738)
0736: Deciphering Complement System-dependent Cellular Pathways in Human Rheumatoid Arthritis Synovial Tissue Using Large Single-cell Computational Omics
Sunday, November 12, 2023
2:45 PM - 2:55 PM PT
Location: Room 32A-B
- FZ
Presenting Author(s)
Juan Vargas1, Ian Mantel2, Nirmal Banda1, Anna Helena Jonsson3, Kevin Wei4, Deepak Rao3, Susan Goodman5, kevin Deane1, The Accelerating Medicines Partnership SLE/RA1, Jennifer Anolik6, Michael Brenner4, Soumya Raychaudhuri3, Accelerating Medicines Partnership (AMP) RA/SLE Network7, Michael Holer1, Laura Donlin5 and Fan Zhang8, 1University of Colorado Anschutz Medical Campus, Aurora, CO, 2Weill Cornell Medicine, New York, NY, 3Brigham and Women's Hospital, Boston, MA, 4Brigham and Women's Hospital and Harvard Medical School, Boston, MA, 5Hospital for Special Surgery, New York, NY, 6University of Rochester Medical Center, Rochester, NY, 7the Accelerating Medicines Partnership (AMP) RA/SLE Network, Boston, MA, 8University of Colorado, Aurora, CO
Background/Purpose: The complement system is a major component of innate immunity and plays a vital role in autoimmune disease pathogenesis. In patients with rheumatoid arthritis (RA), complement activation proteins generated in the synovium can interact with macrophages and other cell types. However, it remains to be determined how complement components directly modulate cell-type functions in the RA synovium to influence phagocytosis and inflammation resolution.
Methods: We have classified the spectrum of RA biopsies into six tissue phenotypes called CTAPs (Cell Type Abundance Phenotypes) using a comprehensive synovial single-cell multimodal atlas ( >314,000 cells, 28 treatment naïve, 27 Methotrexate and 15 TNF inadequate responders) generated from the AMP RA/SLE (1). In this current study, we linked single-cell transcriptomics of complement activation pathway components and receptors with cell-type heterogeneity and tested significant associations with different tissue phenotypes using unsupervised clustering and network analysis. Next, we characterized myeloid cell differentiation using single-cell trajectory analysis and aligned complement gene expression to myeloid functional states. Furthermore, we utilized cell-type interactome analysis to explore whether complement-dependent receptor-ligand pairs are likely to contribute to tissue pathology.
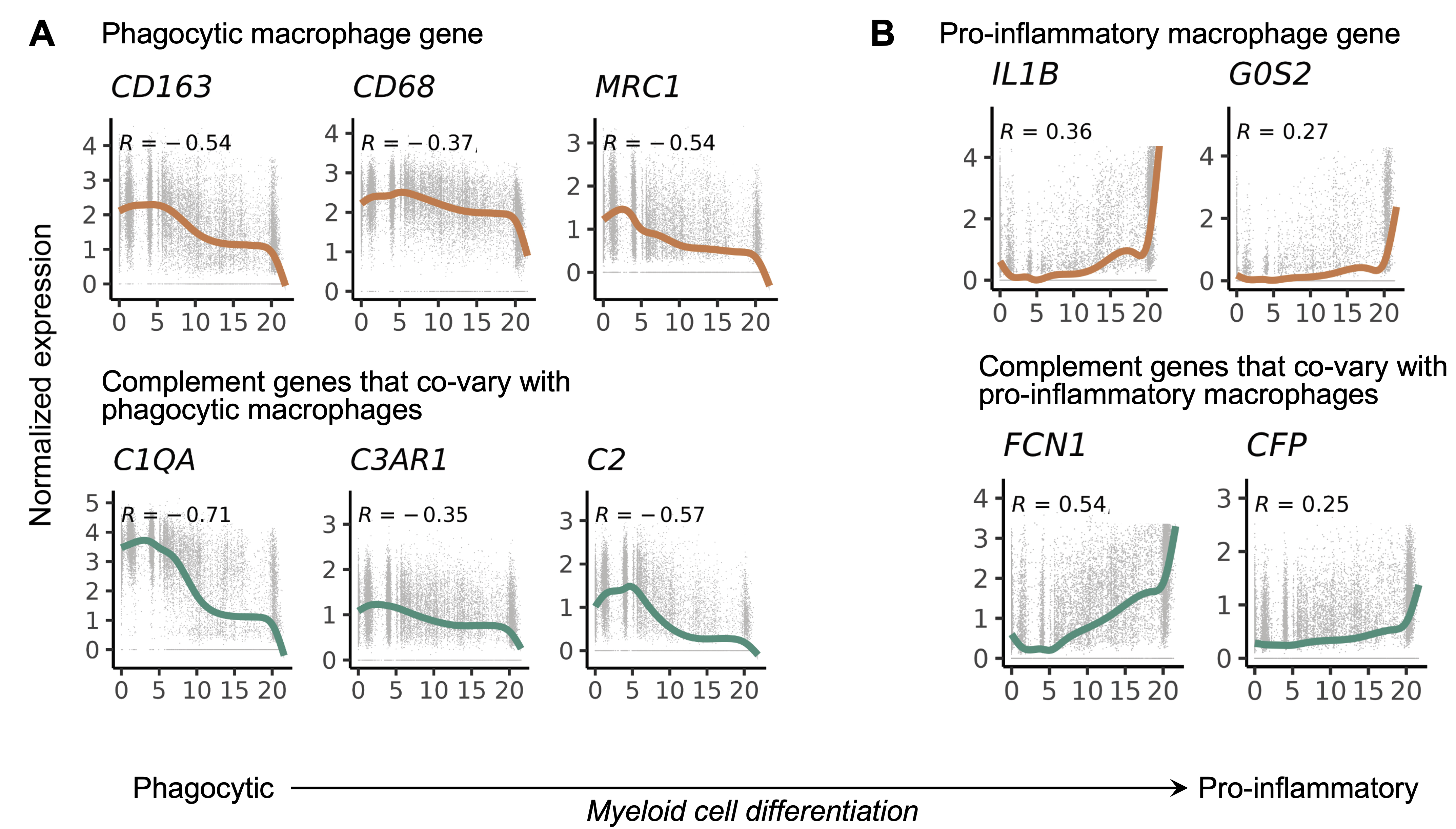
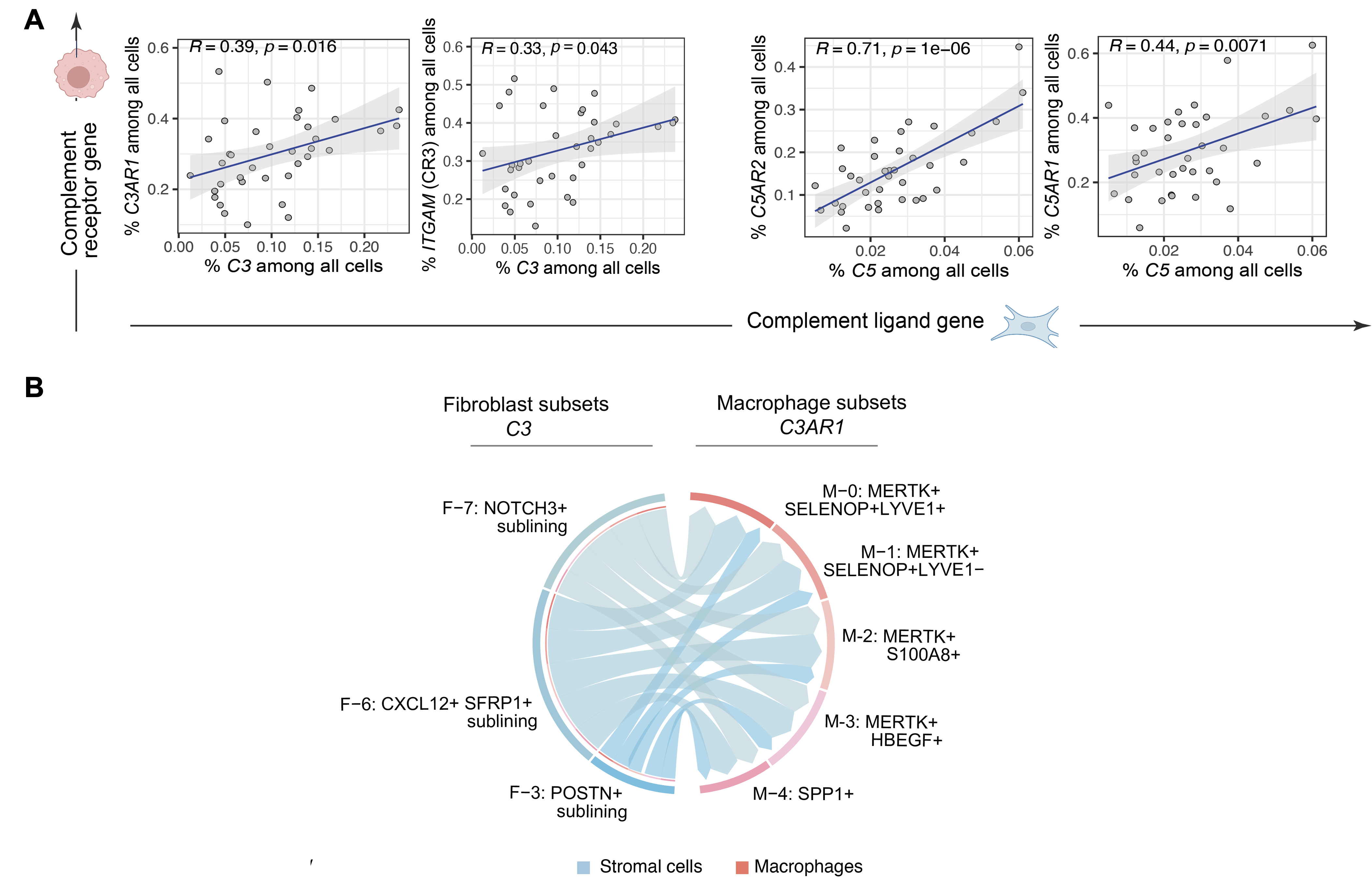
Results: We generated a complement cellular graph characterizing interactions between complement components and cell-type patterns in RA synovium. The complement components present unexpected distinct transcriptomic expression across cell types (Figure 1A). We next systematically captured the relationship between tissue phenotypes with specific complement gene expression (Figure 1B). Within myeloid cells, we identified a myeloid cell differentiation axis revealing that functional macrophage modules align with complement pathways; in particular, complement FCN1 and CFP co-vary with the IL1B+ pro-inflammatory macrophages (p < 2e-16; R=0.48), while complement C1QA-C and C3AR1 co-vary with the MERTK+ phagocytic macrophages (p < 2e-16; R=0.7) (Figure 2). Intriguingly, we revealed that the abundance of complement-dependent receptor-ligand (C3AR1-C3 and C5AR2-C5) are correlated across patients (Figure 3A), and interactome analysis further suggests a potential interaction between the C3AR1 highly expressed MERTK+ macrophages and C3 highly expressed sublining fibroblasts in a particular RA synovial phenotype that relatively lacks lymphocytes (Figure 3B).
Conclusion: Through systematically aligning complement pathways with synovial heterogeneity, we generated a graph describing how complement components can modulate functional cells, especially macrophage phenotypes, in stratified RA patients. This approach of analyzing high-resolution single-cell data in human synovium brings insights into complement pathway-based targets and mechanisms to evaluate in patients with RA, and provides a roadmap for other complex pathways in tissues.
References.
1. Zhang, F. et al. Cellular deconstruction of inflamed synovium defines diverse inflammatory phenotypes in rheumatoid arthritis. bioRxiv 2022 doi:10.1101/2022.02.25.481990.
.jpg)
J. Vargas: None; I. Mantel: None; N. Banda: None; A. Jonsson: None; K. Wei: 10X Genomics, 5, capital one, 6, Gilead sciences, 5, horizon therapeutics, 6, Mestag, 2; D. Rao: AstraZeneca, 2, Bristol-Myers Squibb, 2, 5, GlaxoSmithKlein(GSK), 2, Hifibio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2; S. Goodman: NIH, 5, Novartis, 5; k. Deane: Bristol-Myers Squibb(BMS), 1, Gilead, 5, Janssen, 5, Werfen, 1, 12, Biomarker kits; T. SLE/RA: None; J. Anolik: None; M. Brenner: 4FO Ventures, 2, GlaxoSmithKlein(GSK), 2, Mestag Therapeutics, 2, 11, Third Rock Ventures, 2; S. Raychaudhuri: AbbVie, 6, Janssen, 1, Mestag, Inc, 2, 8, Pfizer, 1, Sanofi, 1, Sonoma, 1, 8; A. Network: None; M. Holer: None; L. Donlin: Bristol-Myers Squibb(BMS), 2, Stryker, 2; F. Zhang: None.
Background/Purpose: The complement system is a major component of innate immunity and plays a vital role in autoimmune disease pathogenesis. In patients with rheumatoid arthritis (RA), complement activation proteins generated in the synovium can interact with macrophages and other cell types. However, it remains to be determined how complement components directly modulate cell-type functions in the RA synovium to influence phagocytosis and inflammation resolution.
Methods: We have classified the spectrum of RA biopsies into six tissue phenotypes called CTAPs (Cell Type Abundance Phenotypes) using a comprehensive synovial single-cell multimodal atlas ( >314,000 cells, 28 treatment naïve, 27 Methotrexate and 15 TNF inadequate responders) generated from the AMP RA/SLE (1). In this current study, we linked single-cell transcriptomics of complement activation pathway components and receptors with cell-type heterogeneity and tested significant associations with different tissue phenotypes using unsupervised clustering and network analysis. Next, we characterized myeloid cell differentiation using single-cell trajectory analysis and aligned complement gene expression to myeloid functional states. Furthermore, we utilized cell-type interactome analysis to explore whether complement-dependent receptor-ligand pairs are likely to contribute to tissue pathology.
Results: We generated a complement cellular graph characterizing interactions between complement components and cell-type patterns in RA synovium. The complement components present unexpected distinct transcriptomic expression across cell types (Figure 1A). We next systematically captured the relationship between tissue phenotypes with specific complement gene expression (Figure 1B). Within myeloid cells, we identified a myeloid cell differentiation axis revealing that functional macrophage modules align with complement pathways; in particular, complement FCN1 and CFP co-vary with the IL1B+ pro-inflammatory macrophages (p < 2e-16; R=0.48), while complement C1QA-C and C3AR1 co-vary with the MERTK+ phagocytic macrophages (p < 2e-16; R=0.7) (Figure 2). Intriguingly, we revealed that the abundance of complement-dependent receptor-ligand (C3AR1-C3 and C5AR2-C5) are correlated across patients (Figure 3A), and interactome analysis further suggests a potential interaction between the C3AR1 highly expressed MERTK+ macrophages and C3 highly expressed sublining fibroblasts in a particular RA synovial phenotype that relatively lacks lymphocytes (Figure 3B).
Conclusion: Through systematically aligning complement pathways with synovial heterogeneity, we generated a graph describing how complement components can modulate functional cells, especially macrophage phenotypes, in stratified RA patients. This approach of analyzing high-resolution single-cell data in human synovium brings insights into complement pathway-based targets and mechanisms to evaluate in patients with RA, and provides a roadmap for other complex pathways in tissues.
References.
1. Zhang, F. et al. Cellular deconstruction of inflamed synovium defines diverse inflammatory phenotypes in rheumatoid arthritis. bioRxiv 2022 doi:10.1101/2022.02.25.481990.
.jpg)
Fig. 1. Single-cell expression distributions of complement components across cell types in rheumatoid arthritis synovial tissues (A). Different myeloid, fibroblast and endothelial cell states express different complement components. Relationship between complement components and RA synovial tissue phenotypes (B).

Fig. 2. Myeloid-specific single-cell trajectory analysis revealed a myeloid cell differentiation trajectory from phagocytic to pro-inflammatory lineage. Complement components C1QA-B, C3AR1, and C2 expressions correlated with anti-inflammatory/phagocytic functions (A); complement components FCN1 and CFP expressions correlated with pro-inflammatory function (B). Spearman correlation R and p-value are shown.

Fig. 3. Across cell type analyses revealed complement-development receptor-ligand correlations (A) and potential complement pathway-dependent C3+ sublininig fibroblasts and MERTK+ tissue macrophages interacting patterns in stratified RA tissues (B). Spearman correlation and p-value are shown.
J. Vargas: None; I. Mantel: None; N. Banda: None; A. Jonsson: None; K. Wei: 10X Genomics, 5, capital one, 6, Gilead sciences, 5, horizon therapeutics, 6, Mestag, 2; D. Rao: AstraZeneca, 2, Bristol-Myers Squibb, 2, 5, GlaxoSmithKlein(GSK), 2, Hifibio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2; S. Goodman: NIH, 5, Novartis, 5; k. Deane: Bristol-Myers Squibb(BMS), 1, Gilead, 5, Janssen, 5, Werfen, 1, 12, Biomarker kits; T. SLE/RA: None; J. Anolik: None; M. Brenner: 4FO Ventures, 2, GlaxoSmithKlein(GSK), 2, Mestag Therapeutics, 2, 11, Third Rock Ventures, 2; S. Raychaudhuri: AbbVie, 6, Janssen, 1, Mestag, Inc, 2, 8, Pfizer, 1, Sanofi, 1, Sonoma, 1, 8; A. Network: None; M. Holer: None; L. Donlin: Bristol-Myers Squibb(BMS), 2, Stryker, 2; F. Zhang: None.



